Latest recommendations

| Id | Title▼ | Authors | Abstract | Picture | Thematic fields | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
02 Jun 2023
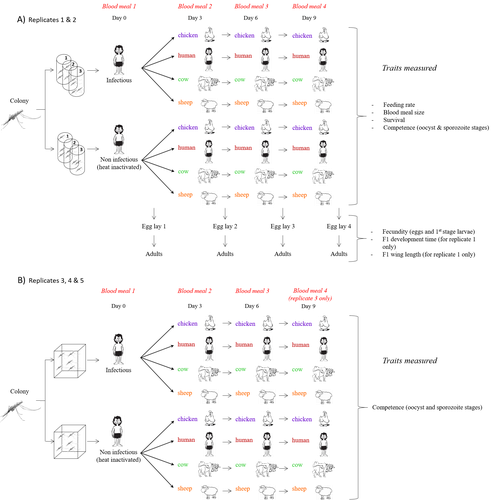
Multiple hosts, multiple impacts: the role of vertebrate host diversity in shaping mosquito life history and pathogen transmissionAmélie Vantaux, Nicolas Moiroux, Kounbobr Roch Dabiré, Anna Cohuet, Thierry Lefèvre https://doi.org/10.1101/2023.02.10.527988What you eat can eliminate you: bloodmeal sources and mosquito fitnessRecommended by Diego Santiago-Alarcon based on reviews by Francisco C. Ferreira and 1 anonymous reviewerDiptera-borne pathogens rank among the most serious health threats to vertebrate organisms around the world, particularly in tropical areas undergoing strong human impacts – e.g., urbanization and farming –, where social unrest and poor economies exacerbate the risk (Allen et al. 2017; Robles-Fernández et al. 2022). Although scientists have acquired a detailed knowledge on the life-history of malaria parasites (Pacheco and Escalante 2023), they still do not have enough information about their insect vectors to make informed management and preventive decisions (Santiago-Alarcon 2022). In this sense, I am pleased to recommend the study of Vantaux et al. (2023), where authors conducted an experimental and theoretical study to analyzed how the diversity of blood sources (i.e., human, cattle, sheep, and chicken) affected the fitness of the human malaria parasite – Plasmodium falciparum – and its mosquito vector – Anopheles gambiae s.l. The study was conducted in Burkina Faso, West Africa. Interestingly, authors did not find a significant effect of blood meal source on parasite development, and a seemingly low impact on the fitness of mosquitoes that were exposed to parasites. However, mosquitoes’ feeding rate, survival, fecundity, and offspring size were negatively affected by the type of blood meal ingested. In general, chicken blood represented the worst meal source for the different measures of mosquito fitness, and sheep blood seems to be the least harmful. This result was supported by the theoretical model, where vectorial capacity was always better when mosquitoes fed on sheep blood compared to cow and chicken blood. Thus, the knowledge generated by this study provides a pathway to reduce human infection risk by managing the diversity of farm animals. For instance, transmission to humans can decrease when chickens and cows represent most of the available blood sources in a village. These results along with other interesting details of this study, represent a clear example of the knowledge and understanding of insect vectors that we need to produce in the future, particularly to manage and prevent hazards and risks (sensu Hoseini et al. 2017). REFERENCES Allen T., et al., Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 8, 1124. (2017). https://doi.org/10.1038/s41467-017-00923-8 Hosseini P.R., et al., Does the impact of biodiversity differ between emerging and endemic pathogens? The need to separate the concepts of hazard and risk. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160129 (2017). https://doi.org/10.1098/rstb.2016.0129 Pacheco M.A., and Escalante, A.A., Origin and diversity of malaria parasites and other Haemosporida. Trend. Parasitol. (2023) https://doi.org/10.1016/j.pt.2023.04.004 Robles-Fernández A., et al., Wildlife susceptibility to infectious diseases at global scales. PNAS 119: e2122851119. (2022). https://doi.org/10.1073/pnas.2122851119 Santiago-Alarcon D. A meta-analytic approach to investigate mosquitoes’ (Diptera: Culicidae) blood feeding preferences from non-urban to urban environments. In: Ecology and Control of Vector-borne Diseases, vol. 7 (R.G. Gutiérrez-López, J.G. Logan, Martínez-de la Puente J., Eds). Pp. 161-177. Wageningen Academic Publishers. eISBN: 978-90-8686-931-2 | ISBN: 978-90-8686-379-2 (2022). Vantaux A. et al. Multiple hosts, multiple impacts: the role of vertebrate host diversity in shaping mosquito life history and pathogen transmission. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community in Infections (2023). https://doi.org/10.1101/2023.02.10.527988 | Multiple hosts, multiple impacts: the role of vertebrate host diversity in shaping mosquito life history and pathogen transmission | Amélie Vantaux, Nicolas Moiroux, Kounbobr Roch Dabiré, Anna Cohuet, Thierry Lefèvre | <p style="text-align: justify;">The transmission of malaria parasites from mosquito to human is largely determined by the dietary specialization of <em>Anopheles mosquitoes</em> to feed on humans. Few studies have explored the impact of blood meal... |  | Ecology of hosts, infectious agents, or vectors, Parasites, Vectors | Diego Santiago-Alarcon | 2023-02-13 11:02:58 | View | |
03 Nov 2023

Longitudinal Survey of Astrovirus infection in different bat species in Zimbabwe: Evidence of high genetic Astrovirus diversityVimbiso Chidoti, Helene De Nys, Malika Abdi, Getrudre Mashura, Valerie Pinarello, Ngoni Chiweshe, Gift Matope, Laure Guerrini, Davies Pfulenyi, Julien Cappelle, Ellen Mwandiringana, Dorothee Misse, Gori Elizabeth, Mathieu Bourgarel, Florian Liegeois https://doi.org/10.1101/2023.04.14.536987High diversity and evidence for inter-species transmission in astroviruses surveyed from bats in ZibabwaeRecommended by Tim James based on reviews by 2 anonymous reviewersMost infectious diseases of humans are zoonoses, and many of these come from particularly species diverse reservoir taxa, such as bats, birds, and rodents (1). Because of our changing landscape, there is increased exposure of humans to wildlife diseases reservoirs, yet we have little basic information about prevalence, hotspots, and transmission factors of most zoonotic pathogens. Viruses are particularly worrisome as a public health risk due to their fast mutation rates and well-known cross-species transmission abilities. There is a global push to better survey wildlife for viruses (2), but these studies are difficult, and the problem is vast. Astroviruses (AstVs) comprise a diverse family of ssRNA viruses known from mammals and birds. Astroviruses can cause gastroenteritis in humans and are more common in elderly and young children, but the relationship of human to non-human Astroviridae as well as transmission routes are unclear. AstVs have been detected at high prevalence in bats in multiple studies (3,4), but it is unclear what factors, such as co-infecting viruses and bat reproductive phenology, influence viral shedding and prevalence. References 1. Mollentze N, Streicker DG. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proceedings of the National Academy of Sciences. 2020 Apr 28;117(17):9423-30. https://doi.org/10.1073/pnas.1919176117 | Longitudinal Survey of Astrovirus infection in different bat species in Zimbabwe: Evidence of high genetic Astrovirus diversity | Vimbiso Chidoti, Helene De Nys, Malika Abdi, Getrudre Mashura, Valerie Pinarello, Ngoni Chiweshe, Gift Matope, Laure Guerrini, Davies Pfulenyi, Julien Cappelle, Ellen Mwandiringana, Dorothee Misse, Gori Elizabeth, Mathieu Bourgarel, Florian Liegeois | <p>Astroviruses (AstVs) have been discovered in over 80 animal species including diverse bat species and avian species. A study on Astrovirus circulation and diversity in different insectivorous and frugivorous chiropteran species roosting in tree... |  | Animal diseases, Epidemiology, Molecular genetics of hosts, infectious agents, or vectors, Reservoirs, Viruses, Zoonoses | Tim James | 2023-04-18 14:58:43 | View | |
28 Sep 2023
Influence of endosymbionts on the reproductive fitness of the tick Ornithodoros moubataTaraveau Florian, Pollet Thomas, Duhayon Maxime, Gardès Laëtitia, Jourdan-Pineau Hélène https://doi.org/10.1101/2023.05.09.539061The cost of endosymbionts on the reproductive fitness of the soft tick Ornithodoros moubataRecommended by Angélique Gobet based on reviews by Luciana Raggi Hoyos and Tuomas Aivelo based on reviews by Luciana Raggi Hoyos and Tuomas Aivelo
Ticks are amongst the most important pathogen vectors in medical and veterinary clinical settings worldwide (Dantas-Torres et al., 2012). Like other holobionts, ticks live in association with a diverse microbiota. It includes tick-borne pathogens (TBP) and other microorganisms that have a beneficial or detrimental effect on the physiology of the host and can also affect the transmission of TBP to animals or humans. In this microbiota, primary endosymbionts, which are obligatory and inheritable, play a role in tick reproduction, the host defense and adaptation to varying environmental conditions (Duron et al., 2018). However, the effect of the microbiota structure and of the endosymbionts on tick fitness and reproduction is not well known. The soft tick Ornithodoros moubata, a parasite known to transmit African swine fever virus (Vial, 2009), is known to host Francisella-like and Rickettsia endosymbionts (Duron et al., 2018). These endosymbionts carry genes involved in B vitamin synthesis which may be supplemented to the host (Bonnet & Pollet, 2021). Here, the authors investigated the role of endosymbionts on the reproductive fitness of Ornithodoros moubata by conducting two experiments (Taraveau et al., 2023). First, they tested the effect of antibiotic treatment of 366 first-stage nymphs on the main endosymbionts Francisella-like and Rickettsia, and measured the endosymbionts presence overtime by qPCR. Second, they surveyed the effect of antibiotic treatment with or without the addition of B vitamins on the survival and reproductive fitness of 132 females over 50 days. This second experiment intended to identify whether the endosymbionts have an effect on the host reproduction or on its nutrition. The supplementation of B vitamin did not have a drastic effect on tick fitness or reproductive traits. However, antibiotic treatments reduced the presence of endosymbionts while increasing tick survival, suggesting a potential cost of hosting endosymbionts on the tick fitness. The authors did a lot of work to thoroughly follow the propositions from Dr Raggi, Dr Aivelo and myself to reconstruct and to revise the manuscript. I believe that the manuscript now reads very well and the answers to the reviews also add some value to the manuscript. As Dr Aivelo pointed out, “this study follows the traditional path of so-called population perturbation studies, where ecologists have administered antibiotics or antihelminths to different animals and seen how the community changes and what effects this has on the host fitness and survival”. As both reviewers stated, results from this study are valuable and provide important basic knowledge that will likely help conduct future experiments on tick microbiota. This recommendation is the result of the thorough reviewing work of Dr Aivelo and Dr Raggi which I warmly thank. Bonnet, S. I., & Pollet, T. (2021). Update on the intricate tango between tick microbiomes and tick‐borne pathogens. Parasite Immunology, 43(5), e12813. https://doi.org/10.1111/pim.12813 Dantas-Torres, F., Chomel, B. B., & Otranto, D. (2012). Ticks and tick-borne diseases: A One Health perspective. Trends in Parasitology, 28(10), 437–446. https://doi.org/10.1016/j.pt.2012.07.003 Duron, O., Morel, O., Noël, V., Buysse, M., Binetruy, F., Lancelot, R., Loire, E., Ménard, C., Bouchez, O., Vavre, F., & Vial, L. (2018). Tick-Bacteria Mutualism Depends on B Vitamin Synthesis Pathways. Current Biology, 28(12), 1896-1902.e5. https://doi.org/10.1016/j.cub.2018.04.038 Taraveau, F., Pollet, T., Duhayon, M., Gardès, L., & Jourdan-Pineau, H. (2023). Influence of endosymbionts on the reproductive fitness of the tick Ornithodoros moubata. bioRxiv, ver.3, peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2023.05.09.539061 Vial, L. (2009). Biological and ecological characteristics of soft ticks (Ixodida: Argasidae) and their impact for predicting tick and associated disease distribution. Parasite, 16(3), 191–202. https://doi.org/10.1051/parasite/2009163191 | Influence of endosymbionts on the reproductive fitness of the tick *Ornithodoros moubata* | Taraveau Florian, Pollet Thomas, Duhayon Maxime, Gardès Laëtitia, Jourdan-Pineau Hélène | <p style="text-align: justify;">Over the past decade, many studies have demonstrated the crucial role of the tick microbiome in tick biology. The soft tick <em>Ornithodoros moubata</em> is a hematophagous ectoparasite of <em>Suidae</em>, best know... | Mutualistic symbionts, Parasites, Pathogenic/Symbiotic Bacteria, Physiology of hosts, infectious agents, or vectors, Vectors | Angélique Gobet | 2023-05-25 19:00:33 | View | ||
07 Oct 2022

Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pestsS. Massart, I. Adams, M. Al Rwahnih, S. Baeyen, G. J. Bilodeau, A. G. Blouin, N. Boonham, T. Candresse, A. Chandelier, K. De Jonghe, A. Fox, Y.Z.A. Gaafar, P. Gentit, A. Haegeman, W. Ho, O. Hurtado-Gonzales, W. Jonkers, J. Kreuze, D. Kutjnak, B. B. Landa, M. Liu, F. Maclot, M. Malapi-Wight, H. J. Maree, F. Martoni, N. Mehle, A. Minafra, D. Mollov, A. G. Moreira, M. Nakhla, F. Petter, A.M. Piper, J. P. Ponchart, R. Rae, B. Remenant, Y. Rivera, B. Rodoni, M. Botermans, J.W. Roenhorst, J. Rollin , ... https://doi.org/10.5281/zenodo.7142136High-throughput sequencing for the diagnostic of plant pathologies and identification of pests: recommendations and challengesRecommended by Olivier Schumpp based on reviews by Denise Altenbach and David RoquisHigh-throughput sequencing (HTS) has revealed an incredible diversity of microorganisms in ecosystems and is also changing the monitoring of macroorganism biodiversity (Deiner et al. 2017; Piper et al. 2019). The diagnostic of plant pathogens and the identification of pests is gradually integrating the use of these techniques, but there are still obstacles. Most of them are related to the reliability of these analyses, which have long been considered insufficient because of their dependence on a succession of sophisticated operations involving parameters that are sometimes difficult to adapt to complex matrices or certain diagnostic contexts. The need to validate HTS approaches is gradually being highlighted in recent work but remains poorly documented (Bester et al. 2022). In this paper, a large community of experts presents and discusses the key steps for optimal control of HTS performance and reliability in a diagnostic context (Massart et al. 2022). It also addresses the issue of costs. The article provides recommendations that closely combine the quality control requirements commonly used in conventional diagnostics with newer or HTS-specific control elements and concepts that are not yet widely used. It discusses the value of these for the use of the various techniques currently covered by the terms "High Throughput Sequencing" in diagnostic activities. The elements presented are intended to limit false positive or false negative results but will also optimise the interpretation of contentious results close to the limits of analytical sensitivity or unexpected results, both of which appear to be frequent when using HTS. Furthermore, the need for risk analysis, verification and validation of methods is well illustrated with numerous examples for each of the steps considered crucial to ensure reliable use of HTS. The clear contextualisation of the proposals made by the authors complements and clarifies the need for user expertise according to the experimental objectives. Some unanswered questions that will require further development and validation are also presented. This article should benefit a large audience including researchers with some level of expertise in HTS but unfamiliar with the recent concepts of controls common in the diagnostic world as well as scientists with strong diagnostic expertise but less at ease with the numerous and complex procedures associated with HTS. References Bester R, Steyn C, Breytenbach JHJ, de Bruyn R, Cook G, Maree HJ (2022) Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids. Plants, 11, 1939. https://doi.org/10.3390/plants11151939 Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872–5895. https://doi.org/10.1111/mec.14350 Massart, S et al. (2022) Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests. Zenodo, 6637519, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6637519 Piper AM, Batovska J, Cogan NOI, Weiss J, Cunningham JP, Rodoni BC, Blacket MJ (2019) Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience, 8, giz092. https://doi.org/10.1093/gigascience/giz092 | Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests | S. Massart, I. Adams, M. Al Rwahnih, S. Baeyen, G. J. Bilodeau, A. G. Blouin, N. Boonham, T. Candresse, A. Chandelier, K. De Jonghe, A. Fox, Y.Z.A. Gaafar, P. Gentit, A. Haegeman, W. Ho, O. Hurtado-Gonzales, W. Jonkers, J. Kreuze, D. Kutjnak, B. B... | <p style="text-align: justify;">High-throughput sequencing (HTS) technologies have the potential to become one of the most significant advances in molecular diagnostics. Their use by researchers to detect and characterize plant pathogens and pests... |  | Diagnosis, Pest management, Phytopathology, Plant diseases | Olivier Schumpp | 2022-06-13 11:26:18 | View | |
06 Apr 2023

Evolution within a given virulence phenotype (pathotype) is driven by changes in aggressiveness: a case study of French wheat leaf rust populationsCécilia FONTYN, Kevin JG MEYER, Anne-Lise BOIXEL, Ghislain DELESTRE, Emma PIAGET, Corentin PICARD, Frédéric SUFFERT, Thierry C MARCEL, Henriette GOYEAU https://doi.org/10.1101/2022.08.29.505401Changes in aggressiveness in pathotypes of wheat leaf rustRecommended by Pierre Gladieux based on reviews by 2 anonymous reviewersUnderstanding the ecological and evolutionary factors underlying the spread of new fungal pathogen populations can inform the development of more effective management strategies. In plant pathology, pathogenicity is generally presented as having two components: ‘virulence’ (qualitative pathogenicity) and aggressiveness (quantitative pathogenicity). Changes in virulence in response to the deployment of new resistant varieties are a major driver of the spread of new populations (called pathotypes, or races) in modern agrosystems, and the genomic (i.e. proximal) and eco-evolutionary (i.e. ultimate) factors underlying these changes are well-documented [1,2,3]. By contrast, the role of changes in aggressiveness in the spread of pathotypes remains little known [4]. The study by Cécilia Fontyn and collaborators [5] set out to characterize changes in aggressiveness for isolates of two pathotypes of the wheat leaf rust (Puccinia triticina) that have been dominant in France during the 2005-2016 period. Isolates were genetically characterized using multilocus microsatellite typing and phenotypically characterized for three components of aggressiveness on wheat varieties: infection efficiency, latency period, and sporulation capacity. Using experiments that represent quite a remarkable amount of work and effort, Fontyn et al. showed that each dominant pathotype consisted of several genotypes, including common genotypes whose frequency changed over time. For each pathotype, the genotypes that were more common initially were replaced by a more aggressive genotype. Together, these results show that changes in the genetic composition of populations of fungal plant pathogens can be associated with, and may be caused by, changes in the quantitative components of pathogenicity. This study also illustrates how extensive, decade-long monitoring of fungal pathogen populations, such as the one conducted for wheat leaf rust in France, represents a very valuable resource for research. REFERENCES [1] Brown, J. K. (1994). Chance and selection in the evolution of barley mildew. Trends in Microbiology, 2(12), 470-475. https://doi.org/10.1016/0966-842x(94)90650-5 [2] Daverdin, G., Rouxel, T., Gout, L., Aubertot, J. N., Fudal, I., Meyer, M., Parlange, F., Carpezat, J., & Balesdent, M. H. (2012). Genome structure and reproductive behaviour influence the evolutionary potential of a fungal phytopathogen. PLoS Pathogens, 8(11), e1003020. https://doi.org/10.1371/journal.ppat.1003020 [3] Gladieux, P., Feurtey, A., Hood, M. E., Snirc, A., Clavel, J., Dutech, C., Roy, M., & Giraud, T. (2015). The population biology of fungal invasions.Molecular Ecology, 24(9), 1969-86. https://doi.org/10.1111/mec.13028 [4] Fontyn, C., Zippert, A. C., Delestre, G., Marcel, T. C., Suffert, F., & Goyeau, H. (2022). Is virulence phenotype evolution driven exclusively by Lr gene deployment in French Puccinia triticina populations?. Plant Pathology, 71(7), 1511-1524. https://doi.org/10.1111/ppa.13599 [5] Fontyn, C., Meyer, K. J., Boixel, A. L., Delestre, G., Piaget, E., Picard, C., Suffer, F., Marcel, T.C., & Goyeau, H. (2022). Evolution within a given virulence phenotype (pathotype) is driven by changes in aggressiveness: a case study of French wheat leaf rust populations. bioRxiv, 2022.08.29.505401, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.08.29.505401 | Evolution within a given virulence phenotype (pathotype) is driven by changes in aggressiveness: a case study of French wheat leaf rust populations | Cécilia FONTYN, Kevin JG MEYER, Anne-Lise BOIXEL, Ghislain DELESTRE, Emma PIAGET, Corentin PICARD, Frédéric SUFFERT, Thierry C MARCEL, Henriette GOYEAU | <p style="text-align: justify;">Plant pathogens are constantly evolving and adapting to their environment, including their host. Virulence alleles emerge, and then increase, and sometimes decrease in frequency within pathogen populations in respon... |  | Coevolution, Epidemiology, Evolution of hosts, infectious agents, or vectors, Interactions between hosts and infectious agents/vectors, Pathogenic/Symbiotic Fungi, Phytopathology, Plant diseases, Population dynamics of hosts, infectious agents, or... | Pierre Gladieux | Emerson Del Ponte , Jacqui Shykoff, Leïla Bagny Beilhe , Alexey Mikaberidze | 2022-09-29 20:01:57 | View |
14 Nov 2022
Ehrlichia ruminantium uses its transmembrane protein Ape to adhere to host bovine aortic endothelial cellsValérie Pinarello, Elena Bencurova, Isabel Marcelino, Olivier Gros, Carinne Puech, Mangesh Bhide, Nathalie Vachiery, Damien F. Meyer https://doi.org/10.1101/2021.06.15.447525Adhesion process of Ehrlichia ruminantium to its host cell: the role of the protein ERGACDS01230 elucidatedRecommended by Thomas Pollet based on reviews by Rodolfo García-Contreras and Alejandro Cabezas-CruzAs recently reported by the world organisation for animal health, 60% of infectious diseases are zoonotic with a significant part associated to ticks. Ticks can transmit various pathogens such as bacteria, viruses and parasites. Among pathogens known to be transmitted by ticks, Ehrlichia ruminantium is an obligate intracellular Gram-negative bacterium responsible for the fatal heartwater disease of domestic and wild ruminants (Allsopp, 2010). E. ruminantium is transmitted by ticks of the genus Amblyomma in the tropical and sub-Saharan areas, as well as in the Caribbean islands. It constitutes a major threat for the American livestock industries since a suitable tick vector is already present in the American mainland and potential introduction of infected A. variegatum through migratory birds or uncontrolled movement of animals from Caribbean could occur (i.e. Deem, 1998 ; Kasari et al 2010). The disease is also a major obstacle to the introduction of animals from heartwater-free to heartwater-infected areas into sub-Saharan Africa and thus restrains breeding programs aiming at upgrading local stocks (Allsopp, 2010). In this context, it is essential to develop control strategies against heartwater, as developing effective vaccines, for instance. Such an objective requires a better understanding of the early interaction of E. ruminantium and its host cells and of the mechanisms associated with bacterial adhesion to the host-cell. In this study, the authors. studied the role of E. ruminantium membrane protein ERGA_CDS_01230 in the adhesion process to host bovine aortic endothelial cells (BAEC). After successfully producing the recombinant version of the protein, Pinarello et al (2022) followed the in vitro culture of E. ruminantium in BAEC and observed that the expression of the protein peaked at the extracellular infectious elementary body stages. This result would suggest the likely involvement of the protein in the early interaction of E. ruminantium with its host cells. The authors then showed using flow cytometry, and scanning electron microscopy, that beads coated with the recombinant protein adhered to BAEC. In addition, they also observed that the adhesion protein of E. ruminantium interacted with proteins of the cell's lysate, membrane and organelle fractions. Additionally, enzymatic treatment, degrading dermatan and chondroitin sulfates on the surface of BAEC, was associated with a 50% reduction in the number of bacteria in the host cell after a development cycle, indicating that glycosaminoglycans might play a role in the adhesion of E. ruminantium to the host-cell. Finally, the authors observed that the adhesion protein of E. ruminantium induced a humoral response in vaccinated animals, making this protein a possible vaccine candidate. As rightly pointed out by both reviewers, the results of this study represent a significant advance (i) in the understanding of the role of the E. ruminantium membrane protein ERGA_CDS_01230 in the adhesion process to the host-cell and (ii) in the development of new control strategies against heartwater as this protein might potentially be used as an immunogen for the development of future vaccines. References Allsopp, B.A. (2010). Natural history of Ehrlichia ruminantium. Vet Parasitol 167, 123-135. https://doi.org/10.1016/j.vetpar.2009.09.014 Deem, S.L. (1998). A review of heartwater and the threat of introduction of Cowdria ruminantium and Amblyomma spp. ticks to the American mainland. J Zoo Wildl Med 29, 109-113. Kasari, T.R. et al (2010). Recognition of the threat of Ehrlichia ruminantium infection in domestic and wild ruminants in the continental United States. J Am Vet Med Assoc. 237:520-30. https://doi.org/10.2460/javma.237.5.520 Pinarello V, Bencurova E, Marcelino I, Gros O, Puech C, Bhide M, Vachiery N, Meyer DF (2022) Ehrlichia ruminantium uses its transmembrane protein Ape to adhere to host bovine aortic endothelial cells. bioRxiv, 2021.06.15.447525, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2021.06.15.447525 | *Ehrlichia ruminantium* uses its transmembrane protein Ape to adhere to host bovine aortic endothelial cells | Valérie Pinarello, Elena Bencurova, Isabel Marcelino, Olivier Gros, Carinne Puech, Mangesh Bhide, Nathalie Vachiery, Damien F. Meyer | <p><em>Ehrlichia ruminantium</em> is an obligate intracellular bacterium, transmitted by ticks of the genus <em>Amblyomma</em> and responsible for heartwater, a disease of domestic and wild ruminants. High genetic diversity of <em>E. ruminantium</... |  | Interactions between hosts and infectious agents/vectors, Microbiology of infections | Thomas Pollet | Rodolfo García-Contreras, Alejandro Cabezas-Cruz | 2021-10-14 16:54:54 | View |
28 Oct 2022

Development of nine microsatellite loci for Trypanosoma lewisi, a potential human pathogen in Western Africa and South-East Asia, and preliminary population genetics analysesAdeline Ségard, Audrey Romero, Sophie Ravel, Philippe Truc, Gauthier Dobigny, Philippe Gauthier, Jonas Etougbetche, Henri-Joel Dossou, Sylvestre Badou, Gualbert Houéménou, Serge Morand, Kittipong Chaisiri, Camille Noûs, Thierry deMeeûs https://doi.org/10.5281/zenodo.6460010Preliminary population genetic analysis of Trypanosoma lewisiRecommended by Annette MacLeod based on reviews by Gabriele Schönian and 1 anonymous reviewerTrypanosoma lewisi is an atypical trypanosome species. Transmitted by fleas, it has a high prevalence and worldwide distribution in small mammals, especially rats [1]. Although not typically thought to infect humans, there has been a number of reports of human infections by T. lewisi in Asia including a case of a fatal infection in an infant [2]. The fact that the parasite is resistant to lysis by normal human serum [3] suggests that many people, especially immunocompromised individuals, may be at risk from zoonotic infections by this pathogen, particularly in regions where there is close contact with T. lewisi-infected rat fleas. Indeed, it is also possible that cryptic T. lewisi infections exist but have hitherto gone undetected. Such asymptomatic infections have been detected for a number of parasitic infections including the related parasite T. b. gambiense [4]. References
[2] Truc P, Büscher P, Cuny G, Gonzatti MI, Jannin J, Joshi P, Juyal P, Lun Z-R, Mattioli R, Pays E, Simarro PP, Teixeira MMG, Touratier L, Vincendeau P, Desquesnes M (2013) Atypical Human Infections by Animal Trypanosomes. PLOS Neglected Tropical Diseases, 7, e2256. https://doi.org/10.1371/journal.pntd.0002256 [3] Lun Z-R, Wen Y-Z, Uzureau P, Lecordier L, Lai D-H, Lan Y-G, Desquesnes M, Geng G-Q, Yang T-B, Zhou W-L, Jannin JG, Simarro PP, Truc P, Vincendeau P, Pays E (2015) Resistance to normal human serum reveals Trypanosoma lewisi as an underestimated human pathogen. Molecular and Biochemical Parasitology, 199, 58–61. https://doi.org/10.1016/j.molbiopara.2015.03.007 [4] Büscher P, Bart J-M, Boelaert M, Bucheton B, Cecchi G, Chitnis N, Courtin D, Figueiredo LM, Franco J-R, Grébaut P, Hasker E, Ilboudo H, Jamonneau V, Koffi M, Lejon V, MacLeod A, Masumu J, Matovu E, Mattioli R, Noyes H, Picado A, Rock KS, Rotureau B, Simo G, Thévenon S, Trindade S, Truc P, Reet NV (2018) Do Cryptic Reservoirs Threaten Gambiense-Sleeping Sickness Elimination? Trends in Parasitology, 34, 197–207. https://doi.org/10.1016/j.pt.2017.11.008 [5] Ségard A, Roméro A, Ravel S, Truc P, Gauthier D, Gauthier P, Dossou H-J, Sylvestre B, Houéménou G, Morand S, Chaisiri K, Noûs C, De Meeûs T (2022) Development of nine microsatellite loci for Trypanosoma lewisi, a potential human pathogen in Western Africa and South-East Asia, and preliminary population genetics analyses. Zenodo, 6460010, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6460010 | Development of nine microsatellite loci for Trypanosoma lewisi, a potential human pathogen in Western Africa and South-East Asia, and preliminary population genetics analyses | Adeline Ségard, Audrey Romero, Sophie Ravel, Philippe Truc, Gauthier Dobigny, Philippe Gauthier, Jonas Etougbetche, Henri-Joel Dossou, Sylvestre Badou, Gualbert Houéménou, Serge Morand, Kittipong Chaisiri, Camille Noûs, Thierry deMeeûs | <p><em>Trypanosoma lewisi</em> belongs to the so-called atypical trypanosomes that occasionally affect humans. It shares the same hosts and flea vector of other medically relevant pathogenic agents as Yersinia pestis, the agent of plague. Increasi... |  | Animal diseases, Disease Ecology/Evolution, Ecology of hosts, infectious agents, or vectors, Eukaryotic pathogens/symbionts, Evolution of hosts, infectious agents, or vectors, Microbiology of infections, Parasites, Population genetics of hosts, in... | Annette MacLeod | 2022-04-21 17:04:37 | View | |
21 Sep 2023
Chikungunya intra-vector dynamics in Aedes albopictus from Lyon (France) upon exposure to a human viremia-like dose range reveals vector barrier permissiveness and supports local epidemic potentialBarbara Viginier, Lucie Cappuccio, Celine Garnier, Edwige Martin, Carine Maisse, Claire Valiente Moro, Guillaume Minard, Albin Fontaine, Sebastian Lequime, Maxime Ratinier, Frederick Arnaud, Vincent Raquin https://doi.org/10.1101/2022.11.06.22281997Fill in one gap in our understanding of CHIKV intra-vector dynamicsRecommended by Sara Moutailler based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
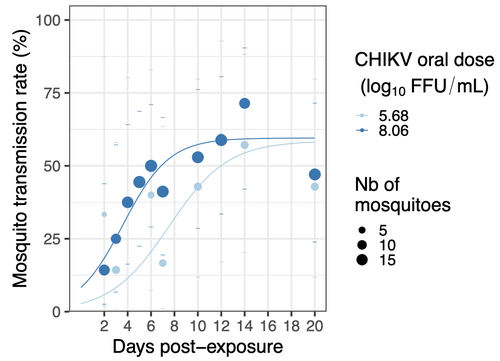
Mosquitoes are first vector of pathogen worldwide and transmit several arbovirus, most of them leading to major outbreaks (1). Chikungunya virus (CHIKV) is a perfect example of the “explosive type” of arbovirus, as observed in La Réunion Island in 2005-2006 (2-6) and also in the outbreak of 2007 in Italy (7), both vectorized by Ae. albopictus. Being able to better understand CHIKV intra-vector dynamics is still of major interest since not all chikungunya strain are explosive ones (8). In this study (9), the authors have evaluated the vector competence of a local strain of Aedes albopictus (collected in Lyon, France) for CHIKV. They evaluated infection, dissemination and transmission dynamics of CHIKV using different dose of virus in individual mosquitoes from day 2 to day 20 post exposure, by titration and quantification of CHIKV RNA load in the saliva. As highlighted by both reviewers, the most innovative idea in this study was the use of three different oral doses trying to span human viraemia detected in two published studies (10-11), doses that were estimated through their model of human CHIKV viremia in the blood. They have found that CHIKV dissemination from the Ae. albopictus midgut depends on the interaction between time post-exposure and virus dose (already highlighted by other international publications). Then their results were implemented in the agent-based model nosoi to estimate the epidemic potential of CHIKV in a French population of Ae. albopictus, using realistic vectorial capacity parameters. To conclude, the authors have discussed the importance of other parameters that could influence vector competence as mosquito microbiota and temperature, parameters that need also to be estimated in local mosquito population to improve the risk assessment through modelling. As pointed out by both reviewers, this is a nice study, well written and easy to read. These results allow filling in another gap of our understanding of CHIKV intra-vector dynamics and highlight the epidemic potential of CHIKV upon transmission by Aedes albopictus in mainland France. For all these reasons, I chose to recommend this article for Peer Community In Infections. References 1. Marine Viglietta, Rachel Bellone, Adrien Albert Blisnick, Anna-Bella Failloux. (2021). Vector Specificity of Arbovirus Transmission. Front Microbiol Dec 9;12:773211. https://doi.org/10.3389/fmicb.2021.773211 2. Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney M-C, Lavenir R, Pardigon N, Reynes J-M, Pettinelli F, Biscornet L, Diancourt L, Michel S, Duquerroy S, Guigon G, Frenkiel M-P, Bréhin A-C, Cubito N, Desprès P, Kunst F, Rey FA, Zeller H, Brisse S. (2006). Genome Microevolution of Chikungunya viruses Causing the Indian Ocean Outbreak. 2006. PLoS Medicine, 3, e263. https://doi.org/10.1371/journal.pmed.0030263 3. Bonilauri P, Bellini R, Calzolari M, Angelini R, Venturi L, Fallacara F, Cordioli P, 687 Angelini P, Venturelli C, Merialdi G, Dottori M. (2008). Chikungunya Virus in Aedes albopictus, Italy. Emerging Infectious 689 Diseases, 14, 852–854. https://doi.org/10.3201/eid1405.071144 4. Pagès F, Peyrefitte CN, Mve MT, Jarjaval F, Brisse S, Iteman I, Gravier P, Tolou H, Nkoghe D, Grandadam M. (2009). Aedes albopictus Mosquito: The Main Vector of the 2007 Chikungunya Outbreak in Gabon. PLoS ONE, 4, e4691. https://doi.org/10.1371/journal.pone.0004691 5. Paupy C, Kassa FK, Caron M, Nkoghé D, Leroy EM (2012) A Chikungunya Outbreak Associated with the Vector Aedes albopictus in Remote Villages of Gabon. Vector-Borne and Zoonotic Diseases, 12, 167–169. https://doi.org/10.1089/vbz.2011.0736 6. Mombouli J-V, Bitsindou P, Elion DOA, Grolla A, Feldmann H, Niama FR, Parra H-J, Munster VJ. (2013). Chikungunya Virus Infection, Brazzaville, Republic of Congo, 2011. Emerging Infectious Diseases, 19, 1542–1543. https://doi.org/10.3201/eid1909.130451 7. Venturi G, Luca MD, Fortuna C, Remoli ME, Riccardo F, Severini F, Toma L, Manso MD, Benedetti E, Caporali MG, Amendola A, Fiorentini C, Liberato CD, Giammattei R, Romi R, Pezzotti P, Rezza G, Rizzo C. (2017). Detection of a chikungunya outbreak in Central Italy, August to September 2017. Eurosurveillance, 22, 17–00646. https://doi.org/10.2807/1560-7917.es.2017.22.39.17-00646 8. de Lima Cavalcanti, T.Y.V.; Pereira, M.R.; de Paula, S.O.; Franca, R.F.d.O. (2022). A Review on Chikungunya Virus Epidemiology, Pathogenesis and Current Vaccine Development. Viruses 2022, 14, 969. https://doi.org/10.3390/v14050969 9. Barbara Viginier, Lucie Cappuccio, Celine Garnier, Edwige Martin, Carine Maisse, Claire Valiente Moro, Guillaume Minard, Albin Fontaine, Sebastian Lequime, Maxime Ratinier, Frederick Arnaud, Vincent Raquin. (2023). Chikungunya intra-vector dynamics in Aedes albopictus from Lyon (France) upon exposure to a human viremia-like dose range reveals vector barrier permissiveness and supports local epidemic potential. medRxiv, ver.3, peer-reviewed and recommended by Peer Community In Infections. https://doi.org/10.1101/2022.11.06.22281997 10. Appassakij H, Khuntikij P, Kemapunmanus M, Wutthanarungsan R, Silpapojakul K (2013) Viremic profiles in CHIKV-infected cases. Transfusion, 53, 2567–2574. https://doi.org/10.1111/j.1537-2995.2012.03960.x 11. Riswari SF, Ma’roef CN, Djauhari H, Kosasih H, Perkasa A, Yudhaputri FA, Artika IM, Williams M, Ven A van der, Myint KS, Alisjahbana B, Ledermann JP, Powers AM, Jaya UA (2015) Study of viremic profile in febrile specimens of chikungunya in Bandung, Indonesia. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology, 74, 61–5. https://doi.org/10.1016/j.jcv.2015.11.017 | Chikungunya intra-vector dynamics in *Aedes albopictus* from Lyon (France) upon exposure to a human viremia-like dose range reveals vector barrier permissiveness and supports local epidemic potential | Barbara Viginier, Lucie Cappuccio, Celine Garnier, Edwige Martin, Carine Maisse, Claire Valiente Moro, Guillaume Minard, Albin Fontaine, Sebastian Lequime, Maxime Ratinier, Frederick Arnaud, Vincent Raquin | <p>Arbovirus emergence and epidemic potential, as approximated by the vectorial capacity formula, depends on host and vector parameters, including the vector intrinsic ability to replicate then transmit the pathogen known as vector competence. Vec... |  | Epidemiology, Vectors, Viruses | Sara Moutailler | 2023-06-17 15:59:17 | View | |
29 Jan 2024
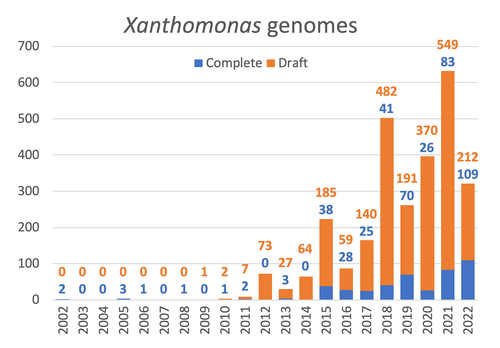
Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy, Provided Insight into the Emergence of Pathogenic Bacteria, Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on XanthomonadsRalf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek https://doi.org/10.5281/zenodo.8223857Advancing Pathogen Genomics: A Comprehensive Review of the Xanthomonas(*) Genome's Impact on Bacterial Research and Control StrategiesRecommended by Damien François Meyer based on reviews by Boris Vinatzer and 3 anonymous reviewers based on reviews by Boris Vinatzer and 3 anonymous reviewers
The paper titled "Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy Provided Insight into the Emergence of Pathogenic Bacteria Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on Xanthomonads" by Ralf Koebnik et al. (2023) is an insightful contribution to the field of genomics and its application in understanding pathogenic bacteria, particularly Xanthomonas. This comprehensive review offers a unique perspective from the French Network on Xanthomonads, underscoring the significant advancements in taxonomy, pathogen emergence, and development of control strategies due to genomic research. One of the paper's main strengths is its thorough exploration of how genomics has revolutionized our understanding of Xanthomonas and other pathogenic bacteria. It sheds light on the evolution and emergence of these pathogens, contributing significantly to the development of novel and effective control measures. The authors' detailed account of the historical progress and current state of genomics in this field highlights its pivotal role in guiding future research and practical applications in managing bacterial diseases. Moreover, the paper emphasizes the importance of collaborative efforts and the sharing of knowledge within scientific networks, as exemplified by the French Network on Xanthomonas. This approach not only enriches the study but also serves as a model for future collaborative research endeavors. In conclusion, the work of Koebnik et al. is a valuable resource for researchers, policymakers, and practitioners in the field of plant pathology and genomics. It not only provides a comprehensive overview of the advances in genomics related to Xanthomonas but also illustrates the broader impact of genomic studies in understanding and managing pathogenic bacteria. References Ralf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek (2024) Celebrating the 20th anniversary of the first Xanthomonas genome gequences – How genomics revolutionized taxonomy, provided insight into the emergence of pathogenic bacteria, enabled new fundamental discoveries and helped developing novel control measures – A perspective from the French network on Xanthomonads. Zenodo ver. 3, peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.8223857 | Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy, Provided Insight into the Emergence of Pathogenic Bacteria, Enabled New Fundamental Discoveries and Helped Developing Novel Control ... | Ralf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek | <p>In this Opinion paper, members of the French Network on Xanthomonads give their personal view on what they consider to be some of the groundbreaking discoveries in the field of molecular plant pathology over the past 20 years. By celebrating th... |  | Epidemiology, Evolution of hosts, infectious agents, or vectors, Genomics, functional genomics of hosts, infectious agents, or vectors, Interactions between hosts and infectious agents/vectors, Molecular biology of infections, Molecular genetics o... | Damien François Meyer | 2023-08-09 10:37:15 | View | |
17 Jan 2024
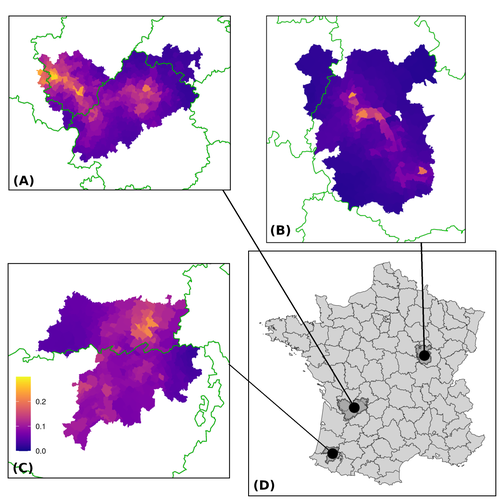
Assessing the dynamics of Mycobacterium bovis infection in three French badger populationsClement CALENGE, Ariane PAYNE, Edouard REVEILLAUD, Celine RICHOMME, Sebastien GIRARD, Stephanie DESVAUX https://doi.org/10.1101/2023.05.31.543041From disease surveillance to public action. Re-inforcing both epidemiological surveillance and data analysis: an illustration with Mycobacterium bovisRecommended by Jean-Francois Guegan based on reviews by Rowland Kao and 1 anonymous reviewerMycobacterium bovis, also called M. tuberculosis var. bovis, is a bacterium belonging to the M. tuberculosis complex (i.e., MTBC) and which can cause through zoonotic transmission another form of human tuberculosis (Tb). It is above all the agent of bovine tuberculosis (i.e., bTb) which affects not only cattle (wild or farmed) but also a large diversity of other wild mammals worldwide. An increasing number of infected animal cases are being discovered in many regions of the world, thus raising the problem of tuberculosis transmission, including to humans, more complex than previously thought. Efforts have been made in terms of vaccination or culling of populations of host carrier species, such as the badger for example, however leading to consequences of greater dispersion of the infectious agent. M. bovis shows a more or less significant capacity to persist outside its hosts, particularly in the environment under certain abiotic and biotic conditions. This bacillus can be transmitted and spread in many ways, including through aerosol, mucus and sputum, urine and feces, by direct contact with infected animals, their dead bodies or rather via their excreta or by inhalation of aerosols, depending on the host species concerned. In this paper, Calenge and his collaborators (Callenge et al. 2024) benefited from a national surveillance program on M. bovis cases in wild species, set up in 2011 in France, i.e., Sylvatub, for detecting and monitoring M. bovis infection in European badger (Meles meles) populations. Sylvatub is a participatory program involving both national and local stakeholder systems in order to determine changes in bTb infection levels in domestic and wild animal species. This original work had two aims: to describe spatial disease dynamics in the three clusters under scrutiny using a complex Bayesian model; and to develop indicators for the monitoring of the M. bovis infection by stakeholders and decision-makers of the program. This paper is timely and very comprehensive. In this cogent study, the authors illustrate this point by using epidemiological surveillance to obtain large amounts of data (which is generally lacking in human epidemiology, but more dramatically lacking in animal epidemiology) and a highly sophisticated biostatistical analysis (Callenge et al. 2024). It is in itself a demonstration of the current capabilities of population dynamics applied to infectious disease situations, in this case animal, in the rapidly developing discipline of disease ecology and evolution. One of the aims of the study is to propose statistical models that can be used by the different stakeholders in charge, for instance, of wildlife conservation or the regional or State veterinary services to assess disease risk in the most affected regions. References Assel AKHMETOVA, Jimena GUERRERO, Paul McADAM, Liliana CM SALVADOR, Joseph CRISPELL, John LAVERY, Eleanor PRESHO, Rowland R KAO, Roman BIEK, Fraser MENZIES, Nigel TRIMBLE, Roland HARWOOD, P Theo PEPLER, Katarina ORAVCOVA, Jordon GRAHAM, Robin SKUCE, Louis DU PLESSIS, Suzan THOMPSON, Lorraine WRIGHT, Andrew W BYRNE, Adrian R ALLEN. 2023. Genomic epidemiology of Mycobacterium bovis infection in sympatric badger and cattle populations in Northern Ireland. Microbial Genomics 9: mgen001023. https://doi.org/10.1099/mgen.0.001023 Roman BIEK, Anthony O’HARE, David WRIGHT, Tom MALLON, Carl McCORMICK, Richard J ORTON, Stanley McDOWELL, Hannah TREWBY, Robin A SKUCE, Rowland R KAO. 2012. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathogens 8: e1003008. https://doi.org/10.1371/journal.ppat.1003008 Clément CALENGE, Ariane PAYNE, Edouard REVEILLAUD, Céline RICHOMME, Sébastien GIRARD, Stephanie DESVAUX. 2024. Assessing the dynamics of Mycobacterium bovis infection in three French badger populations. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community In Infections. https://doi.org/10.1101/2023.05.31.543041 Marc CHOISY, Pejman ROHANI. 2006. Harvesting can increase severity of wildlife disease epidemics. Proceedings of the Royal Society, London, Ser. B 273: 2025-2034. https://doi.org/10.1098/rspb.2006.3554 Shannon C DUFFY, Sreenidhi SRINIVASAN, Megan A SCHILLING, Tod STUBER, Sarah N DANCHUK, Joy S MICHAEL, Manigandan VENKATESAN, Nitish BANSAL, Sushila MAAN, Naresh JINDAL, Deepika CHAUDHARY, Premanshu DANDAPAT, Robab KATANI, Shubhada CHOTHE, Maroudam VEERASAMI, Suelee ROBBE-AUSTERMAN, Nicholas JULEFF, Vivek KAPUR, Marcel A BEHR. 2020. Reconsidering Mycobacterium bovis as a proxy for zoonotic tuberculosis: a molecular epidemiological surveillance study. Lancet Microbe 1: e66-e73. https://doi.org/10.1016/S2666-5247(20)30038-0 Jean-François GUEGAN. 2019. The nature of ecology of infectious disease. The Lancet Infectious Diseases 19. https://doi.org/10.1016/s1473-3099(19)30529-8 Brandon H HAYES, Timothée VERGNE, Mathieu ANDRAUD, Nicolas ROSE. 2023. Mathematical modeling at the livestock-wildlife interface: scoping review of drivers of disease transmission between species. Frontiers in Veterinary Science 10: 1225446. https://doi.org/10.3389/fvets.2023.1225446 David KING, Tim ROPER, Douglas YOUNG, Mark EJ WOOLHOUSE, Dan COLLINS, Paul WOOD. 2007. Bovine tuberculosis in cattle and badgers. Report to Secretary of State about tuberculosis in cattle and badgers. London, UK. https://www.bovinetb.info/docs/RBCT_david_%20king_report.pdf Robert MM SMITH , Francis DROBNIEWSKI, Andrea GIBSON, John DE MONTAGUE, Margaret N LOGAN, David HUNT, Glyn HEWINSON, Roland L SALMON, Brian O’NEILL. 2004. Mycobacterium bovis Infection, United Kingdom. Emerging Infectious Diseases 10: 539-541. https://doi.org/10.3201/eid1003.020819 | Assessing the dynamics of *Mycobacterium bovis* infection in three French badger populations | Clement CALENGE, Ariane PAYNE, Edouard REVEILLAUD, Celine RICHOMME, Sebastien GIRARD, Stephanie DESVAUX | <p>The Sylvatub system is a national surveillance program established in 2011 in France to monitor infections caused by <em>Mycobacterium bovis</em>, the main etiologic agent of bovine tuberculosis, in wild species. This participatory program, inv... |  | Animal diseases, Ecohealth, Ecology of hosts, infectious agents, or vectors, Epidemiology, Geography of infectious diseases, Pathogenic/Symbiotic Bacteria, Zoonoses | Jean-Francois Guegan | 2023-06-05 10:50:49 | View |
MANAGING BOARD
Jorge Amich
Christine Chevillon
Fabrice Courtin
Christine Coustau
Thierry De Meeûs
Heather R. Jordan
Karl-Heinz Kogel
Yannick Moret
Thomas Pollet
Benjamin Roche
Benjamin Rosenthal
Bashir Salim
Lucy Weinert










