
From disease surveillance to public action. Re-inforcing both epidemiological surveillance and data analysis: an illustration with Mycobacterium bovis
 based on reviews by Rowland Kao and 1 anonymous reviewer
based on reviews by Rowland Kao and 1 anonymous reviewer
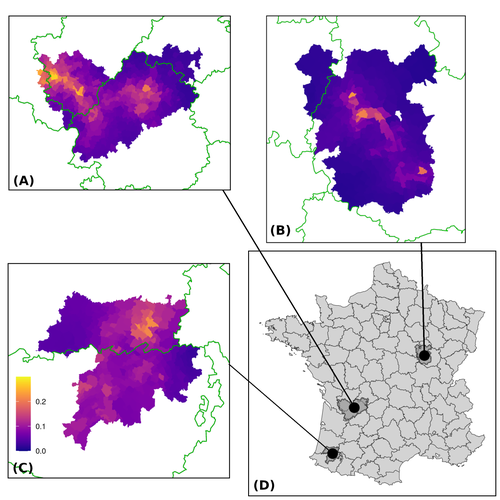
Assessing the dynamics of Mycobacterium bovis infection in three French badger populations
Abstract
Recommendation: posted 17 January 2024, validated 17 January 2024
Guégan, J.-F. (2024) From disease surveillance to public action. Re-inforcing both epidemiological surveillance and data analysis: an illustration with Mycobacterium bovis. Peer Community in Infections, 100088. https://doi.org/10.24072/pci.infections.100088
Recommendation
Mycobacterium bovis, also called M. tuberculosis var. bovis, is a bacterium belonging to the M. tuberculosis complex (i.e., MTBC) and which can cause through zoonotic transmission another form of human tuberculosis (Tb). It is above all the agent of bovine tuberculosis (i.e., bTb) which affects not only cattle (wild or farmed) but also a large diversity of other wild mammals worldwide. An increasing number of infected animal cases are being discovered in many regions of the world, thus raising the problem of tuberculosis transmission, including to humans, more complex than previously thought. Efforts have been made in terms of vaccination or culling of populations of host carrier species, such as the badger for example, however leading to consequences of greater dispersion of the infectious agent. M. bovis shows a more or less significant capacity to persist outside its hosts, particularly in the environment under certain abiotic and biotic conditions. This bacillus can be transmitted and spread in many ways, including through aerosol, mucus and sputum, urine and feces, by direct contact with infected animals, their dead bodies or rather via their excreta or by inhalation of aerosols, depending on the host species concerned.
In this paper, Calenge and his collaborators (Callenge et al. 2024) benefited from a national surveillance program on M. bovis cases in wild species, set up in 2011 in France, i.e., Sylvatub, for detecting and monitoring M. bovis infection in European badger (Meles meles) populations. Sylvatub is a participatory program involving both national and local stakeholder systems in order to determine changes in bTb infection levels in domestic and wild animal species. This original work had two aims: to describe spatial disease dynamics in the three clusters under scrutiny using a complex Bayesian model; and to develop indicators for the monitoring of the M. bovis infection by stakeholders and decision-makers of the program. This paper is timely and very comprehensive.
In this cogent study, the authors illustrate this point by using epidemiological surveillance to obtain large amounts of data (which is generally lacking in human epidemiology, but more dramatically lacking in animal epidemiology) and a highly sophisticated biostatistical analysis (Callenge et al. 2024). It is in itself a demonstration of the current capabilities of population dynamics applied to infectious disease situations, in this case animal, in the rapidly developing discipline of disease ecology and evolution. One of the aims of the study is to propose statistical models that can be used by the different stakeholders in charge, for instance, of wildlife conservation or the regional or State veterinary services to assess disease risk in the most affected regions.
References
Assel AKHMETOVA, Jimena GUERRERO, Paul McADAM, Liliana CM SALVADOR, Joseph CRISPELL, John LAVERY, Eleanor PRESHO, Rowland R KAO, Roman BIEK, Fraser MENZIES, Nigel TRIMBLE, Roland HARWOOD, P Theo PEPLER, Katarina ORAVCOVA, Jordon GRAHAM, Robin SKUCE, Louis DU PLESSIS, Suzan THOMPSON, Lorraine WRIGHT, Andrew W BYRNE, Adrian R ALLEN. 2023. Genomic epidemiology of Mycobacterium bovis infection in sympatric badger and cattle populations in Northern Ireland. Microbial Genomics 9: mgen001023. https://doi.org/10.1099/mgen.0.001023
Roman BIEK, Anthony O’HARE, David WRIGHT, Tom MALLON, Carl McCORMICK, Richard J ORTON, Stanley McDOWELL, Hannah TREWBY, Robin A SKUCE, Rowland R KAO. 2012. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathogens 8: e1003008. https://doi.org/10.1371/journal.ppat.1003008
Clément CALENGE, Ariane PAYNE, Edouard REVEILLAUD, Céline RICHOMME, Sébastien GIRARD, Stephanie DESVAUX. 2024. Assessing the dynamics of Mycobacterium bovis infection in three French badger populations. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community In Infections. https://doi.org/10.1101/2023.05.31.543041
Marc CHOISY, Pejman ROHANI. 2006. Harvesting can increase severity of wildlife disease epidemics. Proceedings of the Royal Society, London, Ser. B 273: 2025-2034. https://doi.org/10.1098/rspb.2006.3554
Shannon C DUFFY, Sreenidhi SRINIVASAN, Megan A SCHILLING, Tod STUBER, Sarah N DANCHUK, Joy S MICHAEL, Manigandan VENKATESAN, Nitish BANSAL, Sushila MAAN, Naresh JINDAL, Deepika CHAUDHARY, Premanshu DANDAPAT, Robab KATANI, Shubhada CHOTHE, Maroudam VEERASAMI, Suelee ROBBE-AUSTERMAN, Nicholas JULEFF, Vivek KAPUR, Marcel A BEHR. 2020. Reconsidering Mycobacterium bovis as a proxy for zoonotic tuberculosis: a molecular epidemiological surveillance study. Lancet Microbe 1: e66-e73. https://doi.org/10.1016/S2666-5247(20)30038-0
Jean-François GUEGAN. 2019. The nature of ecology of infectious disease. The Lancet Infectious Diseases 19. https://doi.org/10.1016/s1473-3099(19)30529-8
Brandon H HAYES, Timothée VERGNE, Mathieu ANDRAUD, Nicolas ROSE. 2023. Mathematical modeling at the livestock-wildlife interface: scoping review of drivers of disease transmission between species. Frontiers in Veterinary Science 10: 1225446. https://doi.org/10.3389/fvets.2023.1225446
David KING, Tim ROPER, Douglas YOUNG, Mark EJ WOOLHOUSE, Dan COLLINS, Paul WOOD. 2007. Bovine tuberculosis in cattle and badgers. Report to Secretary of State about tuberculosis in cattle and badgers. London, UK.
Robert MM SMITH , Francis DROBNIEWSKI, Andrea GIBSON, John DE MONTAGUE, Margaret N LOGAN, David HUNT, Glyn HEWINSON, Roland L SALMON, Brian O’NEILL. 2004. Mycobacterium bovis Infection, United Kingdom. Emerging Infectious Diseases 10: 539-541. https://doi.org/10.3201/eid1003.020819
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Sample collections and analysis were funded by the direction générale de l'alimentation of the French ministry of agriculture.
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2023.05.31.543041
Version of the preprint: 2
Author's Reply, 10 Jan 2024
Decision by Jean-Francois Guégan , posted 30 Oct 2023, validated 30 Oct 2023
, posted 30 Oct 2023, validated 30 Oct 2023
Dear coauthors,
We have now received reports by two reviewers on your work. My apologies for a relatively long review process and due to very many specialists contacted and not responding or not responding quickly. The work is now produced.
As you can read, the two referees are very positive about your work, and suggest a certain number of improvements to increase the quality and understandability of your work. No comment is fatal and you will respond easily. Many of them are requests for details and explanations for easier reading.
I therefore suggest that you respond quickly and point by point, and resubmit a new corrected version; and this in order to be able to recommend your work as quickly as possible.
With many thanks for submitting your work to PCI.
Best regards,
Jean-François Guégan
(Recommender)
Reviewed by anonymous reviewer 1, 04 Jul 2023
Title: “Assessing the dynamics of Mycobacterium bovis infection in three French badger populations”
Authors: Calenge et al.
This paper describes a Bayesian model to analyse the Sylvatub M. bovis surveillance system in order to determine changes in infection levels in badgers in three areas of France. These areas have, in recent years, been the focus of animal tuberculosis control efforts. When risk areas are identified they are subject to different types of surveillance depending on the assigned level of risk. The Sylbatub is used to collect large volumes of data on infection in domestic and wild animal species. In this paper the authors set out to analyse the data to identify indicators that can be easily used to monitor changes in infection.
The paper is very thorough and comprehensive, the authors go into detail explaining the background to the analysis and the development of the analytical model. The outputs of the model allowed for the estimation of prevalence and changes in prevalence with time. A simplified version of the model can be used by stakeholders to assess the risk of infection in the most affected communes.
This paper is quite interesting and highlights the benefits of gathering an interrogating data from infected areas. It moves beyond the simple estimates of point prevalence and changing incidence levels. Where multiple species are implicated in the epidemiology of disease transmission, more sophisticated analysis is required to decipher the key parameters affecting transmission.
Though the manuscript is very detailed there are some areas that could be amended to improve clarity. I do not possess the expertise to critique the details of the Bayesian analysis, though I do have a reasonable understanding of the process as described.
It is not clear, but I do suspect, that the analysis will be sensitive to the trappability of badgers, and we can only assume that it will be similar for infected and non-infected badgers. This could be important as it is stated that only two badgers per commune per year are analysed. One page 3 (Introduction) it mentions that a commune is the smallest division of administration, however, it is on page 6 (materials and methods) that the median size of a commune is defined. I think these values should accompany the statement in the Introduction as it is an obvious question that arises. The TB estimated prevalence in the captured badgers were consistent with the overall bTB rates in the three regions, which was a convincing finding.
Perhaps some comments could be made on the consistency of trapping in the communes across the three regions. This would include the types of traps used (cages?), the number of traps used per commune, the number of trap nights etc.
Among all of the badgers captures in each commune / region, how were the two study animals from each commune chosen?
During necropsy, were the samples collected from each animal based on a pre-selected list of tissues, or did it depend on the lesion status of the animal, i.e., were more issues likely to be collected from an animal presenting with lesions.
A key finding from the study was the low correlation between the infection status of badgers in the same commune was very low. Given that only two badgers were tested in each commune there could be considerable error in this estimate. Although some potential reasons were given, is there any information available on the variation in estimated mean social group size among, for example, neighbouring communes (where they might be predicted to be similar). Do the results provide any information on social group size, density between the regions? The cited paper by Delahay (2000) describes infection in a very high-density population in the UK, which is likely not applicable to most populations (though I acknowledge the point the authors are making here).
Overall, this is a well described study. There are parts where the syntax was not clear and would benefit from a review by a native English language speaker with a good knowledge of this field of research.
Reviewed by Rowland Kao, 25 Oct 2023
This paper is a very useful analysis of French wildlife clusters of bovine TB. I particularly like that the paper addresses both the detailed spatial question and the need for having simpler models accessible to decision-makers and stakeholders. the fact that they compare across all three areas with different patterns of incidence is good and an indicator of robustness. The authors have been very thorough in evaluating the model fit and comparisons however I do have a few suggestions:
i) It would be very useful to have a more extended qualitative description of the data, which would help the reader to understand better what is actually being fitted. The maps themselves are helpful, however temporal and within commune descriptions would also be useful
ii) Line 172 - Not necessary for the study but as a suggestion - the limitation of two tests per community per year seems very low and so I would think this would result in very low confidence in some statistical outcomes– is this true? Of course the authors are limited to the data available to them, however a useful outcome of this study might be an assessment of the utility of increasing this.
iii) Line 183 onwards and figure 2. It’s a bit hard to tell (and in part the reason for the request for a qualitative description of the data) but it looks like the spatial distribution shows very little structure in each area. While I am not an expert in spatial statistics, I do wonder if using random effects per community (as a minor point, unless ‘commune’ is a standardised term I am not aware of, I think ‘community’ would be more usual) may be more than is required to describe the data. It may also result in model overfitting – this would be worth testing.
iv) Line 202. The simulation analysis is welcome and a useful tool. Could the authors better justify the choice of the two scenarios. Also, it might be helpful to identify ways of ‘breaking’ the model – a few possible considerations are greater heterogeneity in spatial risk, and examining the impact of patterns at different stages of an epidemic process – e.g. both early on and later, greater heterogeneities can be observed, depending on the process.
v) Line 243 onwards. The rationale for the simpler approach is clear, and the authors do a good job of demonstrating its utility. However the limitation of no-spatial structure in the model is potentially an important one and so I wonder if in particular, it means that stable spatial structure (even if prevalence is changing, the relative incidence across areas may be stable) is important for this to work (see comment below as well, on proposals for working in new areas and also this reiterates the importance of the qualitative desciption - I can't really tell how the real data are varying in time and space).
vi) Line 486 – the proposal for working with new areas is important if the method is to have broader applicability – a test/train approach could be used to assess the utility here – i.e. how much of a dataset for the complex model is needed to predict the future trends? (To understand the utility of the model for working with new data, it might be worth doing a test/train split (i.e. how good is the correlation between the two models based on a previous model iteration?)
vii) A minor point – the paper overall is to me very clear in the description and the authors should be commended for this. There are however, minor examples throughout of non-standard English usage and it might be useful, for peer review to have someone do a proof read of the manuscript.
https://doi.org/10.24072/pci.infections.100088.rev12









