
SCHUMPP Olivier
- Plant protection, Agroscope, Nyon, Switzerland
- Diagnosis, Phytopathology, Plant diseases, Resistance/Virulence/Tolerance, Taxonomy of hosts, infectious agents, or vectors, Viruses
- recommender
Recommendation: 1
Review: 1
Recommendation: 1

Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests
High-throughput sequencing for the diagnostic of plant pathologies and identification of pests: recommendations and challenges
Recommended by Olivier Schumpp based on reviews by Denise Altenbach and David RoquisHigh-throughput sequencing (HTS) has revealed an incredible diversity of microorganisms in ecosystems and is also changing the monitoring of macroorganism biodiversity (Deiner et al. 2017; Piper et al. 2019).
The diagnostic of plant pathogens and the identification of pests is gradually integrating the use of these techniques, but there are still obstacles. Most of them are related to the reliability of these analyses, which have long been considered insufficient because of their dependence on a succession of sophisticated operations involving parameters that are sometimes difficult to adapt to complex matrices or certain diagnostic contexts. The need to validate HTS approaches is gradually being highlighted in recent work but remains poorly documented (Bester et al. 2022).
In this paper, a large community of experts presents and discusses the key steps for optimal control of HTS performance and reliability in a diagnostic context (Massart et al. 2022). It also addresses the issue of costs. The article provides recommendations that closely combine the quality control requirements commonly used in conventional diagnostics with newer or HTS-specific control elements and concepts that are not yet widely used. It discusses the value of these for the use of the various techniques currently covered by the terms "High Throughput Sequencing" in diagnostic activities. The elements presented are intended to limit false positive or false negative results but will also optimise the interpretation of contentious results close to the limits of analytical sensitivity or unexpected results, both of which appear to be frequent when using HTS.
Furthermore, the need for risk analysis, verification and validation of methods is well illustrated with numerous examples for each of the steps considered crucial to ensure reliable use of HTS. The clear contextualisation of the proposals made by the authors complements and clarifies the need for user expertise according to the experimental objectives. Some unanswered questions that will require further development and validation are also presented.
This article should benefit a large audience including researchers with some level of expertise in HTS but unfamiliar with the recent concepts of controls common in the diagnostic world as well as scientists with strong diagnostic expertise but less at ease with the numerous and complex procedures associated with HTS.
References
Bester R, Steyn C, Breytenbach JHJ, de Bruyn R, Cook G, Maree HJ (2022) Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids. Plants, 11, 1939. https://doi.org/10.3390/plants11151939
Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872–5895. https://doi.org/10.1111/mec.14350
Massart, S et al. (2022) Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests. Zenodo, 6637519, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6637519
Piper AM, Batovska J, Cogan NOI, Weiss J, Cunningham JP, Rodoni BC, Blacket MJ (2019) Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience, 8, giz092. https://doi.org/10.1093/gigascience/giz092
Review: 1
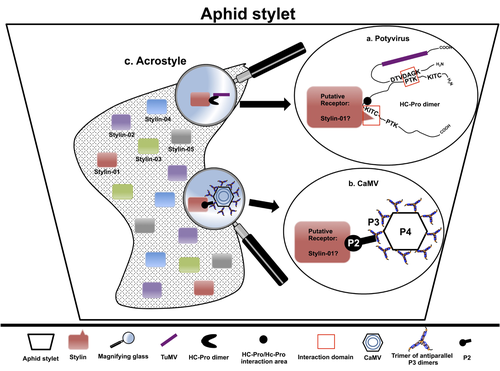
The helper strategy in vector-transmission of plant viruses
The intriguing success of helper components in vector-transmission of plant viruses.
Recommended by Christine Coustau based on reviews by Jamie Bojko and Olivier SchumppMost plant-infecting viruses rely on an animal vector to be transmitted from one sessile host plant to another. A fascinating aspect of virus-vector interactions is the fact that viruses from different clades produce different proteins to bind vector receptors (1). Two major processes are described. In the “capsid strategy”, a motif of the capsid protein is directly binding to the vector receptor. In the “helper strategy”, a non-structural component, the helper component (HC), establishes a bridge between the virus particle and the vector’s receptor.
In this exhaustive review focusing on hemipteran insect vectors, Di Mattia et al. (2) are revisiting the helper strategy in light of recent results. The authors first place the discoveries of the HC strategy in a historical context, suggesting that HC are exclusively found in non-circulative viruses (viruses that only attach to the vector). They present an overview of the nature and modes of action of helper components in the major virus clades of non-circulative viruses (Potyviruses and Caulimoviruses). Authors then detail recent advances, to which they have significantly contributed, showing that the helper strategy also appears widespread in circulative transmission categories (Tenuiviruses, Nanoviruses).
In an extensive perspective section, they raise the question of the evolutionary significance of the existence of HC in numerous unrelated viruses, transmitted by unrelated vectors through different mechanisms. They explore the hypothesis that the helper strategy evolved several times independently in distinct viral clades and for different reasons. In particular, they present several potential benefits of plant virus HC related to virus cooperation, collective transmission and effector-driven infectivity.
As pointed out by both reviewers, this is a very clear and synthetic review. Di Mattia et al. present an exhaustive overview of virus HC-vector molecular interactions and address functionally and evolutionarily important questions. This review should benefit a large audience interested in host-virus interactions and transmission processes.
REFERENCES
(1) Ng JCK, Falk BW (2006) Virus-Vector Interactions Mediating Nonpersistent and Semipersistent Transmission of Plant Viruses. Annual Review of Phytopathology, 44, 183–212. https://doi.org/10.1146/annurev.phyto.44.070505.143325
(2) Di Mattia J, Zeddam J-L, Uzest M, Blanc S (2023) The helper strategy in vector-transmission of plant viruses. Zenodo, ver. 2 peer-reviewed and recommended by Peer Community In Infections. https://doi.org/10.5281/zenodo.7709290