
MEYER Damien François
- Centre for Research and surveillance on Vector-borne diseases in the Caribbean; UMR ASTRE, CIRAD, Petit-Bourg, France
- Animal diseases, Behaviour of hosts, infectious agents, or vectors, Cell biology of hosts, infectious agents, or vectors, Coevolution, Evolution of hosts, infectious agents, or vectors, Interactions between hosts and infectious agents/vectors, Microbiology of infections, Molecular biology of infections, Molecular genetics of hosts, infectious agents, or vectors, Pathogenic/Symbiotic Bacteria, Phylogenetics of hosts, infectious agents, or vectors, Phytopathology, Resistance/Virulence/Tolerance, Vaccines
- recommender
Recommendation: 1
Reviews: 0
Recommendation: 1
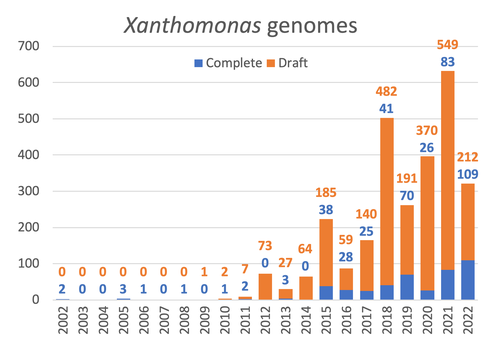
Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy, Provided Insight into the Emergence of Pathogenic Bacteria, Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on Xanthomonads
Advancing Pathogen Genomics: A Comprehensive Review of the Xanthomonas(*) Genome's Impact on Bacterial Research and Control Strategies
Recommended by Damien François Meyer based on reviews by Boris Vinatzer and 3 anonymous reviewersThe paper titled "Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy Provided Insight into the Emergence of Pathogenic Bacteria Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on Xanthomonads" by Ralf Koebnik et al. (2023) is an insightful contribution to the field of genomics and its application in understanding pathogenic bacteria, particularly Xanthomonas. This comprehensive review offers a unique perspective from the French Network on Xanthomonads, underscoring the significant advancements in taxonomy, pathogen emergence, and development of control strategies due to genomic research.
One of the paper's main strengths is its thorough exploration of how genomics has revolutionized our understanding of Xanthomonas and other pathogenic bacteria. It sheds light on the evolution and emergence of these pathogens, contributing significantly to the development of novel and effective control measures. The authors' detailed account of the historical progress and current state of genomics in this field highlights its pivotal role in guiding future research and practical applications in managing bacterial diseases.
Moreover, the paper emphasizes the importance of collaborative efforts and the sharing of knowledge within scientific networks, as exemplified by the French Network on Xanthomonas. This approach not only enriches the study but also serves as a model for future collaborative research endeavors.
In conclusion, the work of Koebnik et al. is a valuable resource for researchers, policymakers, and practitioners in the field of plant pathology and genomics. It not only provides a comprehensive overview of the advances in genomics related to Xanthomonas but also illustrates the broader impact of genomic studies in understanding and managing pathogenic bacteria.
References
Ralf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek (2024) Celebrating the 20th anniversary of the first Xanthomonas genome gequences – How genomics revolutionized taxonomy, provided insight into the emergence of pathogenic bacteria, enabled new fundamental discoveries and helped developing novel control measures – A perspective from the French network on Xanthomonads. Zenodo ver. 3, peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.8223857