Latest recommendations

| Id | Title * | Authors * | Abstract * | Picture * | Thematic fields * | Recommender | Reviewers▲ | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
08 Dec 2022
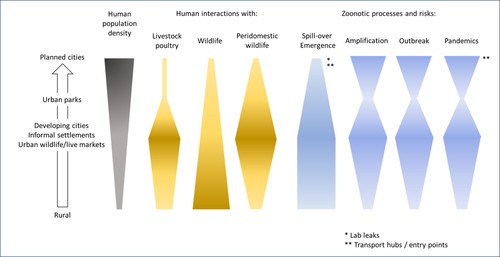
Zoonotic emergence at the animal-environment-human interface: the forgotten urban socio-ecosystemsDobigny, G. & Morand, S. https://doi.org/10.5281/zenodo.6444776Zoonotic emergence and the overlooked case of citiesRecommended by Etienne Waleckx based on reviews by Eric Dumonteil, Nicole L. Gottdenker and 1 anonymous reviewerZoonotic pathogens, those transmitted from animals to humans, constitute a major public health risk with high associated global economic costs. Diseases associated with these pathogens represent more than 60% of emerging infectious diseases and predominantly originate in wildlife (1). Over the last decades, the emergence and re-emergence of zoonotic pathogens have led to an increasing number of epidemics, as illustrated by the current Covid-19 pandemic. There is ample evidence that human impact on native ecosystems such as deforestation, agricultural development, and urbanization, is linked to spillover of pathogens from animals to humans (2). However, research and calls to action have mainly focused on the importance of surveillance and prevention of zoonotic emergences along landscape interfaces, with special emphasis on tropical forests and agroecosystems, and studies and reviews pointing out the zoonotic risk associated with cities are scarce. Additionally, cities are sometimes wrongly seen as one homogeneous ecosystem, almost exclusively human, with a Northern hemisphere-biased perception of what a city is, which fails to take into account the ecological and socio-economic diversities that can constitute an urban area. Here, Dobigny and Morand (3) aim to draw attention to the importance of urban ecosystems in zoonotic risk and advocate that further attention should be paid to urban, peri-urban and suburban areas. In this well-organized and well-documented review, the authors show, using updated literature, that cities are places where massive contacts occur between wildlife, domestic animals, and human inhabitants (thus constituting spillover opportunities), and that it is even likely that human and wildlife contact in urban centers is more prevalent than in wild areas, perhaps contrary to intuition. Indeed, cities currently constitute the most important environment of human life and are places for millions of close interactions between humans and animals, including pets and domestic animals, wild animals through the intrusion of wild urban-adapted species (e.g., some bat, rodent, or bird species among others), manipulation and consumption of wildlife meat, and the existence of wildlife meat markets, which all constitute a major risk for zoonotic spillover. In cities, lab escapees of zoonotic pathogens also exist, and trends of adaptation to urban ecological conditions of many vectors of primary health importance is also a concern. The authors further argue that cities are predominant places for both epidemic amplification of human-human transmitted pathogens, because they are places with high human densities and population growth, and for dissemination of reservoirs, vectors and pathogens, as they are transport hubs. Dobigny & Morand further predict, likely correctly, that cities may be important places for pathogen evolution. Finally, they propose actions and recommendations to limit the risk of zoonotic spillover events from urban ecosystems and future directions for research aiming at assessing this risk. The reviewers found the manuscript well-organized and presented, timely, and bringing novel contributions to the field of zoonotic emergence. I strongly recommend this article, which should benefit a large audience, particularly in the context of the current Covid-19 pandemics and the ongoing One Health initiatives aiming at preventing future zoonotic disease emergence (4). References (1) Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P (2008) Global trends in emerging infectious diseases. Nature, 451, 990–993. https://doi.org/10.1038/nature06536 (2) White RJ, Razgour O (2020) Emerging zoonotic diseases originating in mammals: a systematic review of effects of anthropogenic land-use change. Mammal Review, 50, 336–352. https://doi.org/10.1111/mam.12201 (3) Dobigny G, Morand S (2022) Zoonotic emergence at the animal-environment-human interface: the forgotten urban socio-ecosystems. Zenodo, 6444776, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6444776 (4) Morand S, Lajaunie C (2021) Biodiversity and COVID-19: A report and a long road ahead to avoid another pandemic. One Earth, 4, 920–923. https://doi.org/10.1016/j.oneear.2021.06.007 | Zoonotic emergence at the animal-environment-human interface: the forgotten urban socio-ecosystems | Dobigny, G. & Morand, S. | <p style="text-align: justify;">Zoonotic emergence requires spillover from animals to humans, hence animal-human interactions. A lot has been emphasized on human intrusion into wild habitats (e.g., deforestation, hunting) and the development of ag... |  | Disease Ecology/Evolution, Ecohealth, Ecology of hosts, infectious agents, or vectors, Evolution of hosts, infectious agents, or vectors, One Health, Zoonoses | Etienne Waleckx | 2022-04-11 11:39:11 | View | |
28 Oct 2022

Development of nine microsatellite loci for Trypanosoma lewisi, a potential human pathogen in Western Africa and South-East Asia, and preliminary population genetics analysesAdeline Ségard, Audrey Romero, Sophie Ravel, Philippe Truc, Gauthier Dobigny, Philippe Gauthier, Jonas Etougbetche, Henri-Joel Dossou, Sylvestre Badou, Gualbert Houéménou, Serge Morand, Kittipong Chaisiri, Camille Noûs, Thierry deMeeûs https://doi.org/10.5281/zenodo.6460010Preliminary population genetic analysis of Trypanosoma lewisiRecommended by Annette MacLeod based on reviews by Gabriele Schönian and 1 anonymous reviewerTrypanosoma lewisi is an atypical trypanosome species. Transmitted by fleas, it has a high prevalence and worldwide distribution in small mammals, especially rats [1]. Although not typically thought to infect humans, there has been a number of reports of human infections by T. lewisi in Asia including a case of a fatal infection in an infant [2]. The fact that the parasite is resistant to lysis by normal human serum [3] suggests that many people, especially immunocompromised individuals, may be at risk from zoonotic infections by this pathogen, particularly in regions where there is close contact with T. lewisi-infected rat fleas. Indeed, it is also possible that cryptic T. lewisi infections exist but have hitherto gone undetected. Such asymptomatic infections have been detected for a number of parasitic infections including the related parasite T. b. gambiense [4]. References
[2] Truc P, Büscher P, Cuny G, Gonzatti MI, Jannin J, Joshi P, Juyal P, Lun Z-R, Mattioli R, Pays E, Simarro PP, Teixeira MMG, Touratier L, Vincendeau P, Desquesnes M (2013) Atypical Human Infections by Animal Trypanosomes. PLOS Neglected Tropical Diseases, 7, e2256. https://doi.org/10.1371/journal.pntd.0002256 [3] Lun Z-R, Wen Y-Z, Uzureau P, Lecordier L, Lai D-H, Lan Y-G, Desquesnes M, Geng G-Q, Yang T-B, Zhou W-L, Jannin JG, Simarro PP, Truc P, Vincendeau P, Pays E (2015) Resistance to normal human serum reveals Trypanosoma lewisi as an underestimated human pathogen. Molecular and Biochemical Parasitology, 199, 58–61. https://doi.org/10.1016/j.molbiopara.2015.03.007 [4] Büscher P, Bart J-M, Boelaert M, Bucheton B, Cecchi G, Chitnis N, Courtin D, Figueiredo LM, Franco J-R, Grébaut P, Hasker E, Ilboudo H, Jamonneau V, Koffi M, Lejon V, MacLeod A, Masumu J, Matovu E, Mattioli R, Noyes H, Picado A, Rock KS, Rotureau B, Simo G, Thévenon S, Trindade S, Truc P, Reet NV (2018) Do Cryptic Reservoirs Threaten Gambiense-Sleeping Sickness Elimination? Trends in Parasitology, 34, 197–207. https://doi.org/10.1016/j.pt.2017.11.008 [5] Ségard A, Roméro A, Ravel S, Truc P, Gauthier D, Gauthier P, Dossou H-J, Sylvestre B, Houéménou G, Morand S, Chaisiri K, Noûs C, De Meeûs T (2022) Development of nine microsatellite loci for Trypanosoma lewisi, a potential human pathogen in Western Africa and South-East Asia, and preliminary population genetics analyses. Zenodo, 6460010, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6460010 | Development of nine microsatellite loci for Trypanosoma lewisi, a potential human pathogen in Western Africa and South-East Asia, and preliminary population genetics analyses | Adeline Ségard, Audrey Romero, Sophie Ravel, Philippe Truc, Gauthier Dobigny, Philippe Gauthier, Jonas Etougbetche, Henri-Joel Dossou, Sylvestre Badou, Gualbert Houéménou, Serge Morand, Kittipong Chaisiri, Camille Noûs, Thierry deMeeûs | <p><em>Trypanosoma lewisi</em> belongs to the so-called atypical trypanosomes that occasionally affect humans. It shares the same hosts and flea vector of other medically relevant pathogenic agents as Yersinia pestis, the agent of plague. Increasi... |  | Animal diseases, Disease Ecology/Evolution, Ecology of hosts, infectious agents, or vectors, Eukaryotic pathogens/symbionts, Evolution of hosts, infectious agents, or vectors, Microbiology of infections, Parasites, Population genetics of hosts, in... | Annette MacLeod | 2022-04-21 17:04:37 | View | |
07 Feb 2023
Three-way relationships between gut microbiota, helminth assemblages and bacterial infections in wild rodent populationsMarie Bouilloud, Maxime Galan, Adelaide Dubois, Christophe Diagne, Philippe Marianneau, Benjamin Roche, Nathalie Charbonnel https://doi.org/10.1101/2022.05.23.493084Unveiling the complex interactions between members of gut microbiomes: a significant advance provided by an exhaustive study of wild bank volesRecommended by Thomas Pollet based on reviews by Jason Anders and 1 anonymous reviewerThe gut of vertebrates is a host for hundreds or thousands of different species of microorganisms named the gut microbiome. This latter may differ greatly in natural environments between individuals, populations and species (1). The vertebrate gut microbiome plays key roles in host fitness through functions including nutrient acquisition, immunity and defense against infectious agents. While bank voles are small mammals potentially reservoirs of a large number of infectious agents, questions about the links between their gut microbiome and the presence of pathogens are scarcely addressed. In this study, Bouilloud et al. (2) used complementary analyses of community and microbial ecology to (i) assess the variability of gut bacteriome diversity and composition in wild populations of the bank vole Myodes glareolus collected in four different sites in Eastern France and (ii) evaluate the three-way interactions between the gut bacteriota, the gastro-intestinal helminths and pathogenic bacteria detected in the spleen. Authors identified important variations of the gut bacteriota composition and diversity among bank voles mainly explained by sampling localities. They found positive correlations between the specific richness of both the gut bacteria and the helminth community, as well as between the composition of these two communities, even when accounting for the influence of geographical distance. The helminths Aonchotheca murissylvatici, Heligmosomum mixtum and the bacteria Bartonella sp were the main taxa associated with the whole gut bacteria composition. Besides, changes in relative abundance of particular gut bacterial taxa were specifically associated with other helminths (Mastophorus muris, Catenotaenia henttoneni, Paranoplocephala omphalodes and Trichuris arvicolae) or pathogenic bacteria. Infections with Neoehrlichia mikurensis, Orientia sp, Rickettsia sp and P. omphalodes were especially associated with lower relative abundance of members of the family Erysipelotrichaceae (Firmicutes), while coinfections with higher number of bacterial infections were associated with lower relative abundance of members of the Bacteroidales family (Bacteroidetes). As pointed out by both reviewers, this study represents a significant advance in the field. I would like to commend the authors for this enormous work. The amount of data, analyses and results is considerable which has sometimes complicated the understanding of the story at the beginning of the evaluation process. Thanks to constructive scientific interactions with both reviewers through the two rounds of evaluation, the authors have efficiently addressed the reviewer's concerns and improved the manuscript, making this great story easier to read. The innovative results of this study emphasize the complex interlinkages between gut bacteriome and infections in wild animal populations and I strongly recommend this article for publication In Peer Community Infections. References (1) Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y (2020) Host variables confound gut microbiota studies of human disease. Nature, 587, 448–454. https://doi.org/10.1038/s41586-020-2881-9 (2) Bouilloud M, Galan M, Dubois A, Diagne C, Marianneau P, Roche B, Charbonnel N (2023) Three-way relationships between gut microbiota, helminth assemblages and bacterial infections in wild rodent populations. biorxiv, 2022.05.23.493084, ver. 2 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.05.23.493084 | Three-way relationships between gut microbiota, helminth assemblages and bacterial infections in wild rodent populations | Marie Bouilloud, Maxime Galan, Adelaide Dubois, Christophe Diagne, Philippe Marianneau, Benjamin Roche, Nathalie Charbonnel | <p>Background</p> <p>Despite its central role in host fitness, the gut microbiota may differ greatly between individuals. This variability is often mediated by environmental or host factors such as diet, genetics, and infections. Recently, a part... | Disease Ecology/Evolution, Ecohealth, Interactions between hosts and infectious agents/vectors, Reservoirs, Zoonoses | Thomas Pollet | 2022-05-25 10:13:23 | View | ||
07 Oct 2022

Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pestsS. Massart, I. Adams, M. Al Rwahnih, S. Baeyen, G. J. Bilodeau, A. G. Blouin, N. Boonham, T. Candresse, A. Chandelier, K. De Jonghe, A. Fox, Y.Z.A. Gaafar, P. Gentit, A. Haegeman, W. Ho, O. Hurtado-Gonzales, W. Jonkers, J. Kreuze, D. Kutjnak, B. B. Landa, M. Liu, F. Maclot, M. Malapi-Wight, H. J. Maree, F. Martoni, N. Mehle, A. Minafra, D. Mollov, A. G. Moreira, M. Nakhla, F. Petter, A.M. Piper, J. P. Ponchart, R. Rae, B. Remenant, Y. Rivera, B. Rodoni, M. Botermans, J.W. Roenhorst, J. Rollin , ... https://doi.org/10.5281/zenodo.7142136High-throughput sequencing for the diagnostic of plant pathologies and identification of pests: recommendations and challengesRecommended by Olivier Schumpp based on reviews by Denise Altenbach and David RoquisHigh-throughput sequencing (HTS) has revealed an incredible diversity of microorganisms in ecosystems and is also changing the monitoring of macroorganism biodiversity (Deiner et al. 2017; Piper et al. 2019). The diagnostic of plant pathogens and the identification of pests is gradually integrating the use of these techniques, but there are still obstacles. Most of them are related to the reliability of these analyses, which have long been considered insufficient because of their dependence on a succession of sophisticated operations involving parameters that are sometimes difficult to adapt to complex matrices or certain diagnostic contexts. The need to validate HTS approaches is gradually being highlighted in recent work but remains poorly documented (Bester et al. 2022). In this paper, a large community of experts presents and discusses the key steps for optimal control of HTS performance and reliability in a diagnostic context (Massart et al. 2022). It also addresses the issue of costs. The article provides recommendations that closely combine the quality control requirements commonly used in conventional diagnostics with newer or HTS-specific control elements and concepts that are not yet widely used. It discusses the value of these for the use of the various techniques currently covered by the terms "High Throughput Sequencing" in diagnostic activities. The elements presented are intended to limit false positive or false negative results but will also optimise the interpretation of contentious results close to the limits of analytical sensitivity or unexpected results, both of which appear to be frequent when using HTS. Furthermore, the need for risk analysis, verification and validation of methods is well illustrated with numerous examples for each of the steps considered crucial to ensure reliable use of HTS. The clear contextualisation of the proposals made by the authors complements and clarifies the need for user expertise according to the experimental objectives. Some unanswered questions that will require further development and validation are also presented. This article should benefit a large audience including researchers with some level of expertise in HTS but unfamiliar with the recent concepts of controls common in the diagnostic world as well as scientists with strong diagnostic expertise but less at ease with the numerous and complex procedures associated with HTS. References Bester R, Steyn C, Breytenbach JHJ, de Bruyn R, Cook G, Maree HJ (2022) Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids. Plants, 11, 1939. https://doi.org/10.3390/plants11151939 Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872–5895. https://doi.org/10.1111/mec.14350 Massart, S et al. (2022) Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests. Zenodo, 6637519, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6637519 Piper AM, Batovska J, Cogan NOI, Weiss J, Cunningham JP, Rodoni BC, Blacket MJ (2019) Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience, 8, giz092. https://doi.org/10.1093/gigascience/giz092 | Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests | S. Massart, I. Adams, M. Al Rwahnih, S. Baeyen, G. J. Bilodeau, A. G. Blouin, N. Boonham, T. Candresse, A. Chandelier, K. De Jonghe, A. Fox, Y.Z.A. Gaafar, P. Gentit, A. Haegeman, W. Ho, O. Hurtado-Gonzales, W. Jonkers, J. Kreuze, D. Kutjnak, B. B... | <p style="text-align: justify;">High-throughput sequencing (HTS) technologies have the potential to become one of the most significant advances in molecular diagnostics. Their use by researchers to detect and characterize plant pathogens and pests... |  | Diagnosis, Pest management, Phytopathology, Plant diseases | Olivier Schumpp | 2022-06-13 11:26:18 | View | |
14 Dec 2022

Transcriptome responses of the aphid vector Myzus persicae are shaped by identities of the host plant and the virusQuentin Chesnais, Victor Golyaev, Amandine Velt, Camille Rustenholz, Maxime Verdier, Véronique Brault, Mikhail M. Pooggin, Martin Drucker https://doi.org/10.1101/2022.07.18.500449How do multiple host plants and virus species challenge aphid molecular machinery?Recommended by Sebastien Massart based on reviews by Juan José Lopez Moya and Michelle HeckThe impact of virus infection of a plant on an aphid’s behaviour has been observed in many studies [1]. Indeed, virus infection can alter plant biochemistry through the emission of volatile organic compounds and plant tissue content modification. These alterations can further impact the interactions between plants and aphids. However, although it is a well-known phenomenon, very few studies have explored the consequences of plant virus infection on the gene expression of aphids to understand better the aphid’s manipulation by the plant virus. In this context, the recommended study [2] reports a comprehensive transcriptomic analysis of the genes expressed by one aphid species, Myzus persicae, a vector of several plant viruses, when feeding on plants. Michelle Heck underlined how significant this study is for comprehending the molecular bases of aphid-vector manipulation by plant viruses (see below). Interestingly, the study design has integrated several factors that might influence the gene expression of M. persicae when feeding on the plant. Indeed, the authors investigated the effect of two plant species (Arabidopsis thaliana and Camelia sativa) and two virus species [turnip yellows virus (TuYV) and cauliflower mosaic virus (CaMV)]. Noteworthy, the transmission mode of TuYV is circulative and persistent, while CaMV is transmitted by a semi-persistent non-circulative mode. As Juan José Lopez Moya mentioned, multiple comparisons allowed the identification of the different responses of aphids in front of different host plants infected or not by different viruses (see below). This publication is complementary to a previous publication from the same team focusing on plant transcriptome analysis [3]. Thanks to their experimental design, the authors identified genes commonly deregulated by both viruses and/or both plant species and deregulated genes by a single virus or a single plant. Figure 4 nicely summarizes the number of deregulated genes. A thorough discussion on the putative role of deregulated genes in different conditions gave a comprehensive follow-up of the results and their impact on the current knowledge of plant-virus-vector interactions. This study has now opened the gate to promising research focusing on the functional validation of the identified genes while also narrowing the study from the body to the tissue level. References: 1. Carr JP, Tungadi T, Donnelly R, Bravo-Cazar A, Rhee S-J, Watt LG, Mutuku JM, Wamonje FO, Murphy AM, Arinaitwe W, Pate AE, Cunniffe NJ, Gilligan CA (2020) Modelling and manipulation of aphid-mediated spread of non-persistently transmitted viruses. Virus Research, 277, 197845. https://doi.org/10.1016/j.virusres.2019.197845 2. Chesnais Q, Golyaev V, Velt A, Rustenholz C, Verdier M, Brault V, Pooggin MM, Drucker M (2022) Transcriptome responses of the aphid vector Myzus persicae are shaped by identities of the host plant and the virus. bioRxiv , 2022.07.18.500449, ver. 5 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.07.18.500449 3. Chesnais Q, Golyaev V, Velt A, Rustenholz C, Brault V, Pooggin MM, Drucker M (2022) Comparative Plant Transcriptome Profiling of Arabidopsis thaliana Col-0 and Camelina sativa var. Celine Infested with Myzus persicae Aphids Acquiring Circulative and Noncirculative Viruses Reveals Virus- and Plant-Specific Alterations Relevant to Aphid Feeding Behavior and Transmission. Microbiology Spectrum, 10, e00136-22. https://doi.org/10.1128/spectrum.00136-22 | Transcriptome responses of the aphid vector *Myzus persicae* are shaped by identities of the host plant and the virus | Quentin Chesnais, Victor Golyaev, Amandine Velt, Camille Rustenholz, Maxime Verdier, Véronique Brault, Mikhail M. Pooggin, Martin Drucker | <p style="text-align: justify;"><strong>Background:</strong> Numerous studies have documented modifications in vector orientation behavior, settling and feeding behavior, and/or fecundity and survival due to virus infection in host plants. These a... |  | Behaviour of hosts, infectious agents, or vectors, Cell biology of hosts, infectious agents, or vectors, Molecular biology of infections, Physiology of hosts, infectious agents, or vectors, Phytopathology, Plant diseases, Vectors, Viruses | Sebastien Massart | 2022-07-19 15:24:14 | View | |
23 Mar 2023
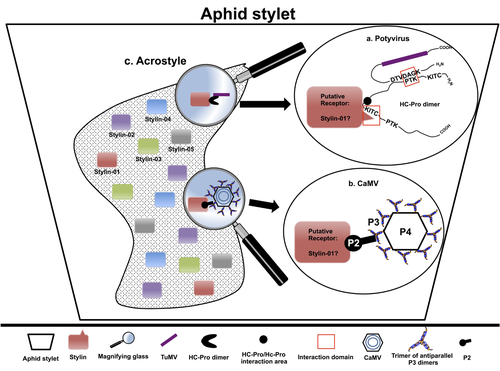
The helper strategy in vector-transmission of plant virusesDi Mattia Jérémy, Zeddam Jean Louis, Uzest Marilyne and Stéphane Blanc https://doi.org/10.5281/zenodo.7709290The intriguing success of helper components in vector-transmission of plant viruses.Recommended by Christine Coustau based on reviews by Jamie Bojko and Olivier SchumppMost plant-infecting viruses rely on an animal vector to be transmitted from one sessile host plant to another. A fascinating aspect of virus-vector interactions is the fact that viruses from different clades produce different proteins to bind vector receptors (1). Two major processes are described. In the “capsid strategy”, a motif of the capsid protein is directly binding to the vector receptor. In the “helper strategy”, a non-structural component, the helper component (HC), establishes a bridge between the virus particle and the vector’s receptor. In this exhaustive review focusing on hemipteran insect vectors, Di Mattia et al. (2) are revisiting the helper strategy in light of recent results. The authors first place the discoveries of the HC strategy in a historical context, suggesting that HC are exclusively found in non-circulative viruses (viruses that only attach to the vector). They present an overview of the nature and modes of action of helper components in the major virus clades of non-circulative viruses (Potyviruses and Caulimoviruses). Authors then detail recent advances, to which they have significantly contributed, showing that the helper strategy also appears widespread in circulative transmission categories (Tenuiviruses, Nanoviruses). In an extensive perspective section, they raise the question of the evolutionary significance of the existence of HC in numerous unrelated viruses, transmitted by unrelated vectors through different mechanisms. They explore the hypothesis that the helper strategy evolved several times independently in distinct viral clades and for different reasons. In particular, they present several potential benefits of plant virus HC related to virus cooperation, collective transmission and effector-driven infectivity. As pointed out by both reviewers, this is a very clear and synthetic review. Di Mattia et al. present an exhaustive overview of virus HC-vector molecular interactions and address functionally and evolutionarily important questions. This review should benefit a large audience interested in host-virus interactions and transmission processes. REFERENCES (1) Ng JCK, Falk BW (2006) Virus-Vector Interactions Mediating Nonpersistent and Semipersistent Transmission of Plant Viruses. Annual Review of Phytopathology, 44, 183–212. https://doi.org/10.1146/annurev.phyto.44.070505.143325 (2) Di Mattia J, Zeddam J-L, Uzest M, Blanc S (2023) The helper strategy in vector-transmission of plant viruses. Zenodo, ver. 2 peer-reviewed and recommended by Peer Community In Infections. https://doi.org/10.5281/zenodo.7709290 | The helper strategy in vector-transmission of plant viruses | Di Mattia Jérémy, Zeddam Jean Louis, Uzest Marilyne and Stéphane Blanc | <p>An intriguing aspect of vector-transmission of plant viruses is the frequent involvement of a helper component (HC). HCs are virus-encoded non-structural proteins produced in infected plant cells that are mandatory for the transmission success.... |  | Evolution of hosts, infectious agents, or vectors, Interactions between hosts and infectious agents/vectors, Molecular biology of infections, Molecular genetics of hosts, infectious agents, or vectors, Plant diseases, Vectors, Viruses | Christine Coustau | 2022-10-28 17:32:39 | View | |
25 Apr 2023

The distribution, phenology, host range and pathogen prevalence of Ixodes ricinus in France: a systematic map and narrative reviewGrégoire Perez, Laure Bournez, Nathalie Boulanger, Johanna Fite, Barbara Livoreil, Karen D. McCoy, Elsa Quillery, Magalie René-Martellet, and Sarah I. Bonnet https://doi.org/10.1101/2023.04.18.537315An extensive review of Ixodes ricinus in European FranceRecommended by Ana Sofia Santos based on reviews by Ana Palomar and 1 anonymous reviewerTicks are obligate, bloodsucking, nonpermanent ectoparasitic arthropods. Among them, Ixodes ricinus is a classic example of an extreme generalist tick, presenting a highly permissive feeding behavior using different groups of vertebrates as hosts, such as mammalian (including humans), avian and reptilian species (Hoogstraal & Aeschlimann, 1982; Dantas-Torresa & Otranto, 2013). This ecological adaptation can account for the broad geographical distribution of I. ricinus populations, which extends from the western end of the European continent to the Ural Mountains in Russia, and from northern Norway to the Mediterranean basin, including the North African countries - Morocco, Algeria and Tunisia (https://ecdc.europa.eu/en/disease-vectors/surveillance-and-disease-data/tick-maps). The contact with different hosts also promotes the exposure/acquisition and transmission of various pathogenic agents (viruses, bacteriae, protists and nematodes) of veterinary and medical relevance (Aeschlimann et al., 1979). As one of the prime ticks found on humans, this species is implicated in diseases such as Lyme borreliosis, Spotted Fever Group rickettsiosis, Human Anaplasmosis, Human Babesiosis and Tick-borne Encephalitis (Velez et al., 2023). The climate change projections drawn for I. ricinus, in the scenario of global warming, point for the expansion/increase activity in both latitude and altitude (Medlock et al., 2013). The adequacy of vector modeling is relaying in the proper characterization of complex biological systems. Thus, it is essential to increase knowledge on I. ricinus, focusing on aspects such as genetic background, ecology and eco-epidemiology on a microscale but also at a country and region level, due to possible local adaptations of tick populations and genetic drift. In the present systematic revision, Perez et al. (2023) combine old and recently published data (mostly up to 2020) regarding I. ricinus distribution, phenology, host range and pathogen association in continental France and Corsica Island. Based on a keyword search of peer-reviewed papers on seven databases, as well as other sources of grey literature (mostly, thesis), the authors have synthesized information on: 1) Host parasitism to detect potential differences in host use comparing to other areas in Europe; 2) The spatiotemporal distribution of I. ricinus, to identify possible geographic trends in tick density, variation in activity patterns and the influence of environmental factors; 3) Tick-borne pathogens detected in this species, to better assess their spatial distribution and variation in exposure risk. As pointed out by both reviewers, this work clearly summarizes the information regarding I. ricinus and associated microorganisms from European France. This review also identifies remaining knowledge gaps, providing a comparable basis to orient future research. This is why I chose to recommend Perez et al (2023)'s preprint for Peer Community Infections. REFERENCES Aeschlimann, A., Burgdorfer, W., Matile, H., Peter, O., Wyler, R. (1979) Aspects nouveaux du rôle de vecteur joué par Ixodes ricinus L. en Suisse. Acta Tropica, 36, 181-191. Dantas-Torresa, F., Otranto, D. (2013) Seasonal dynamics of Ixodes ricinus on ground level and higher vegetation in a preserved wooded area in southern Europe. Veterinary Parasitology, 192, 253- 258. Hoogstraal, H., Aeschlimann, A. (1982) Tick-host specificity. Mitteilungen der Schweizerischen Entomologischen Gesellschaft, 55, 5-32. Medlock, J.M., Hansford, K.M., Bormane, A., Derdakova, M., Estrada-Peña, A., George, J.C., Golovljova, I., Jaenson, T.G.T., Jensen, J.K., Jensen, P.M., Kazimirova, M., Oteo, J.A., Papa, A., Pfister, K., Plantard, O., Randolph, S.E., Rizzoli, A., Santos-Silva, M.M., Sprong, H., Vial, L., Hendrickx, G., Zeller, H., Van Bortel, W. (2013) Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasites and Vectors, 6. https://doi.org/10.1186/1756-3305-6-1 Perez, G., Bournez, L., Boulanger, N., Fite, J., Livoreil, B., McCoy, K., Quillery, E., René-Martellet, M., Bonnet, S. (2023) The distribution, phenology, host range and pathogen prevalence of Ixodes ricinus in France: a systematic map and narrative review. bioRxiv, ver. 1 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2023.04.18.537315 Velez, R., De Meeûs, T., Beati, L., Younsi, H., Zhioua, E., Antunes, S., Domingos, A., Ataíde Sampaio, D., Carpinteiro, D., Moerbeck, L., Estrada-Peña, A., Santos-Silva, M.M., Santos, A.S. (2023) Development and testing of microsatellite loci for the study of population genetics of Ixodes ricinus Linnaeus, 1758 and Ixodes inopinatus Estrada-Peña, Nava & Petney, 2014 (Acari: Ixodidae) in the western Mediterranean region. Acarologia, 63, 356-372. https://doi.org/10.24349/bvem-4h49 | The distribution, phenology, host range and pathogen prevalence of *Ixodes ricinus* in France: a systematic map and narrative review | Grégoire Perez, Laure Bournez, Nathalie Boulanger, Johanna Fite, Barbara Livoreil, Karen D. McCoy, Elsa Quillery, Magalie René-Martellet, and Sarah I. Bonnet | <p style="text-align: justify;">The tick <em>Ixodes ricinus</em> is the most important vector species of infectious diseases in European France. Understanding its distribution, phenology, and host species use, along with the distribution and preva... |  | Animal diseases, Behaviour of hosts, infectious agents, or vectors, Disease Ecology/Evolution, Ecohealth, Ecology of hosts, infectious agents, or vectors, Epidemiology, Geography of infectious diseases, Interactions between hosts and infectious ag... | Ana Sofia Santos | 2022-12-06 14:52:44 | View | |
27 Feb 2023
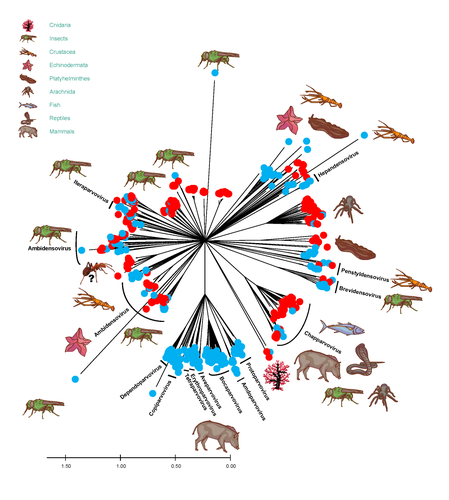
African army ants at the forefront of virome surveillance in a remote tropical forestMatthieu Fritz, Berenice Reggiardo, Denis Filloux, Lisa Claude, Emmanuel Fernandez, Frederic Mahe, Simona Kraberger, Joy M. Custer, Pierre Becquart, Telstar Ndong Mebaley, Linda Bohou Kombila, Leadisaelle H. Lenguiya, Larson Boundenga, Illich M. Mombo, Gael Darren Maganga, Fabien R. Niama, Jean-Sylvain Koumba, Mylene Ogliastro, Michel Yvon, Darren Martin, Stephane Blanc, Arvind Varsani, Eric Leroy, Philippe Roumagnac https://doi.org/10.1101/2022.12.13.520061A groundbreaking study using ants revealed a spectacular diversity of viruses in hardly accessible ecosystems like tropical forestsRecommended by Sebastien Massart based on reviews by Mart Krupovic and 1 anonymous reviewerDeciphering the virome (the set or assemblage of viruses) of the Earth, from individual organisms to entire ecosystems, has become a key priority. The first step to better understanding the impact of viruses on the ecology and functions of ecosystems is to describe their diversity. Such knowledge opens the gates to a better assessment of global nutrient cycling or of the threat that viruses represent to individual health. This explains the increasing number of pioneering studies that are currently sequencing the complete or partial genome of thousands of new viruses [1]. In their exciting study, Fritz and collaborators [2], authors sampled 209 army ants (Genus Dorylus) to investigate the virus diversity in dense forests that researchers cannot easily access. Indeed, these ants live in colonies (21 were sampled) that can move 1 km per day, covering a significant area and attacking many invertebrate and vertebrate preys. Each sample was sequenced by a protocol called VANA sequencing and allowing the enrichment of the sample in viral sequences [3], so improving the detection of viruses present at low abundance in the ant (and more specifically in its gut for viruses infecting preys). Around 45,000 contigs presented homologies with bacterial, plant, invertebrate, and vertebrate infecting viruses. Half could be assigned to 56 families and 157 genera of the International Committee on Taxonomy of Viruses. Beyond this amazing harvest of new and known virus sequences using an original methodology, the results significantly improve the current frontiers of known viral taxonomy and diversity and raise exciting research tracks to expand them. As a preprint, several blogs or news of leading scientists and journals have already highlighted this study. For example, in the news section of Science magazine, Jon Cohen underlined the originality of the approach for virus hunting on Earth with the title “Armed with air samplers, rope tricks, and—yes—ants, virus hunters spot threats in new ways”[4]. Another example is the mention of the publication by Elisabeth Bik in her Microbiome Digest: she wrote, “An amazing read is a fresh preprint from Fritz and collaborator describing an exciting method of sampling in difficult-to-reach environments“ [5]. The paper from Fritz et al [2] thus represents a significant advance in virus ecology, as already recognized by early readers, and this is why I strongly recommend its publication in PCI Infections. REFERENCES 1. Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D, Lohr D, Novakovsky G, Buchfink B, Al-Shayeb B, Banfield JF, de la Peña M, Korobeynikov A, Chikhi R, Babaian A (2022) Petabase-scale sequence alignment catalyses viral discovery. Nature, 602, 142–147. https://doi.org/10.1038/s41586-021-04332-2 2. Fritz M, Reggiardo B, Filloux D, Claude L, Fernandez E, Mahé F, Kraberger S, Custer JM, Becquart P, Mebaley TN, Kombila LB, Lenguiya LH, Boundenga L, Mombo IM, Maganga GD, Niama FR, Koumba J-S, Ogliastro M, Yvon M, Martin DP, Blanc S, Varsani A, Leroy E, Roumagnac P (2023) African army ants at the forefront of virome surveillance in a remote tropical forest. bioRxiv, 2022.12.13.520061, ver. 4 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.12.13.520061 3. François S, Filloux D, Fernandez E, Ogliastro M, Roumagnac P (2018) Viral Metagenomics Approaches for High-Resolution Screening of Multiplexed Arthropod and Plant Viral Communities. In: Viral Metagenomics: Methods and Protocols Methods in Molecular Biology. (eds Pantaleo V, Chiumenti M), pp. 77–95. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-7683-6_7 4. Cohen J (2023) Virus hunters test new surveillance tools. Science, 379, 16–17. https://doi.org/10.1126/science.adg5292 5. Ponsero A (2023) February 18th, 2023. Microbiome Digest - Bik’s Picks. https://microbiomedigest.com/2023/02/18/february-18th-2023/ | African army ants at the forefront of virome surveillance in a remote tropical forest | Matthieu Fritz, Berenice Reggiardo, Denis Filloux, Lisa Claude, Emmanuel Fernandez, Frederic Mahe, Simona Kraberger, Joy M. Custer, Pierre Becquart, Telstar Ndong Mebaley, Linda Bohou Kombila, Leadisaelle H. Lenguiya, Larson Boundenga, Illich M. M... | <p style="text-align: justify;">In this study, we used a predator-enabled metagenomics strategy to sample the virome of a remote and difficult-to-access densely forested African tropical region. Specifically, we focused our study on the use of arm... |  | Ecohealth, Ecology of hosts, infectious agents, or vectors, One Health, Reservoirs, Viruses | Sebastien Massart | 2022-12-14 11:57:40 | View | |
02 Jun 2023
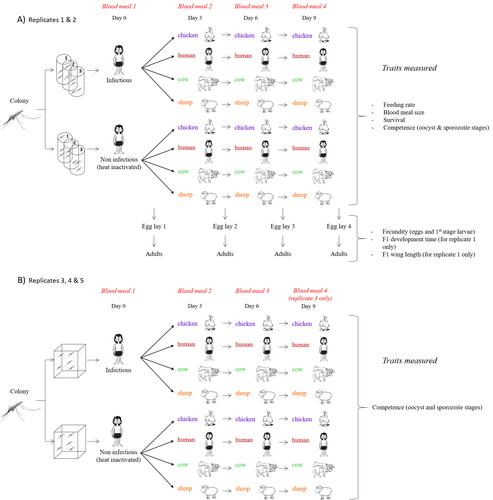
Multiple hosts, multiple impacts: the role of vertebrate host diversity in shaping mosquito life history and pathogen transmissionAmélie Vantaux, Nicolas Moiroux, Kounbobr Roch Dabiré, Anna Cohuet, Thierry Lefèvre https://doi.org/10.1101/2023.02.10.527988What you eat can eliminate you: bloodmeal sources and mosquito fitnessRecommended by Diego Santiago-Alarcon based on reviews by Francisco C. Ferreira and 1 anonymous reviewerDiptera-borne pathogens rank among the most serious health threats to vertebrate organisms around the world, particularly in tropical areas undergoing strong human impacts – e.g., urbanization and farming –, where social unrest and poor economies exacerbate the risk (Allen et al. 2017; Robles-Fernández et al. 2022). Although scientists have acquired a detailed knowledge on the life-history of malaria parasites (Pacheco and Escalante 2023), they still do not have enough information about their insect vectors to make informed management and preventive decisions (Santiago-Alarcon 2022). In this sense, I am pleased to recommend the study of Vantaux et al. (2023), where authors conducted an experimental and theoretical study to analyzed how the diversity of blood sources (i.e., human, cattle, sheep, and chicken) affected the fitness of the human malaria parasite – Plasmodium falciparum – and its mosquito vector – Anopheles gambiae s.l. The study was conducted in Burkina Faso, West Africa. Interestingly, authors did not find a significant effect of blood meal source on parasite development, and a seemingly low impact on the fitness of mosquitoes that were exposed to parasites. However, mosquitoes’ feeding rate, survival, fecundity, and offspring size were negatively affected by the type of blood meal ingested. In general, chicken blood represented the worst meal source for the different measures of mosquito fitness, and sheep blood seems to be the least harmful. This result was supported by the theoretical model, where vectorial capacity was always better when mosquitoes fed on sheep blood compared to cow and chicken blood. Thus, the knowledge generated by this study provides a pathway to reduce human infection risk by managing the diversity of farm animals. For instance, transmission to humans can decrease when chickens and cows represent most of the available blood sources in a village. These results along with other interesting details of this study, represent a clear example of the knowledge and understanding of insect vectors that we need to produce in the future, particularly to manage and prevent hazards and risks (sensu Hoseini et al. 2017). REFERENCES Allen T., et al., Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 8, 1124. (2017). https://doi.org/10.1038/s41467-017-00923-8 Hosseini P.R., et al., Does the impact of biodiversity differ between emerging and endemic pathogens? The need to separate the concepts of hazard and risk. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160129 (2017). https://doi.org/10.1098/rstb.2016.0129 Pacheco M.A., and Escalante, A.A., Origin and diversity of malaria parasites and other Haemosporida. Trend. Parasitol. (2023) https://doi.org/10.1016/j.pt.2023.04.004 Robles-Fernández A., et al., Wildlife susceptibility to infectious diseases at global scales. PNAS 119: e2122851119. (2022). https://doi.org/10.1073/pnas.2122851119 Santiago-Alarcon D. A meta-analytic approach to investigate mosquitoes’ (Diptera: Culicidae) blood feeding preferences from non-urban to urban environments. In: Ecology and Control of Vector-borne Diseases, vol. 7 (R.G. Gutiérrez-López, J.G. Logan, Martínez-de la Puente J., Eds). Pp. 161-177. Wageningen Academic Publishers. eISBN: 978-90-8686-931-2 | ISBN: 978-90-8686-379-2 (2022). Vantaux A. et al. Multiple hosts, multiple impacts: the role of vertebrate host diversity in shaping mosquito life history and pathogen transmission. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community in Infections (2023). https://doi.org/10.1101/2023.02.10.527988 | Multiple hosts, multiple impacts: the role of vertebrate host diversity in shaping mosquito life history and pathogen transmission | Amélie Vantaux, Nicolas Moiroux, Kounbobr Roch Dabiré, Anna Cohuet, Thierry Lefèvre | <p style="text-align: justify;">The transmission of malaria parasites from mosquito to human is largely determined by the dietary specialization of <em>Anopheles mosquitoes</em> to feed on humans. Few studies have explored the impact of blood meal... |  | Ecology of hosts, infectious agents, or vectors, Parasites, Vectors | Diego Santiago-Alarcon | 2023-02-13 11:02:58 | View | |
19 Jul 2023
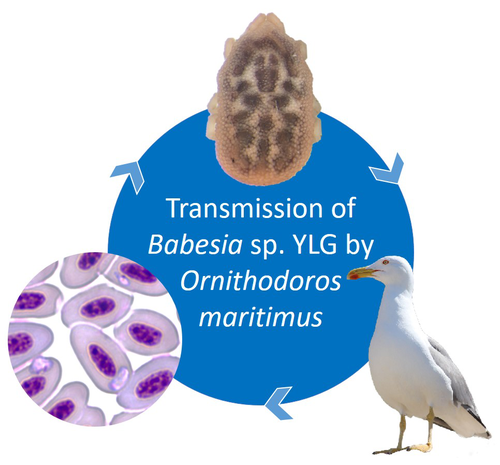
A soft tick vector of Babesia sp. YLG in Yellow-legged gull (Larus michahellis) nestsClaire Bonsergent, Marion Vittecoq, Carole Leray, Maggy Jouglin, Marie Buysse, Karen D. McCoy, Laurence Malandrin https://doi.org/10.1101/2023.03.24.534071A four-year study reveals the potential role of the soft tick Ornithodoros maritimus in the transmission and circulation of Babesia sp. YLG in Yellow-legged gull colonies.Recommended by Thomas Pollet based on reviews by Hélène Jourdan-Pineau and Tahar KernifWorldwide, ticks and tick-borne diseases are a persistent example of problems at the One Health interface between humans, wildlife, and environment (1, 2). The management and prevention of ticks and tick-borne diseases require a better understanding of host, tick and pathogen interactions and thus get a better view of the tick-borne pathosystems. In this study (3), the tick-borne pathosystem included three component species: first a seabird host, the Yellow-legged gull (YLG - Larus michahellis, Laridae), second a soft nidicolous tick (Ornithodoros maritimus, Argasidae, syn. Alectorobius maritimus) known to infest this host and third a blood parasite (Babesia sp. YLG, Piroplasmidae). In this pathosystem, authors investigated the role of the soft tick, Ornithodoros maritimus, as a potential vector of Babesia sp. YLG. They analyzed the transmission of Babesia sp. YLG by collecting different tick life stages from YLG nests during 4 consecutive years on the islet of Carteau (Gulf of Fos, Camargue, France). Ticks were dissected and organs were analyzed separately to detect the presence of Babesia sp DNA and to evaluate different transmission pathways. While the authors detected Babesia sp. YLG DNA in the salivary glands of nymphs, females and males, this result reveals a strong suspicion of transmission of the parasite by the soft tick. Babesia sp. YLG DNA was also found in tick ovaries, which could indicate possible transovarial transmission. Finally, the authors detected Babesia sp. YLG DNA in several male testes and in endospermatophores, and notably in a parasite-free female (uninfected ovaries and salivary glands). These last results raise the interesting possibility of sexual transmission from infected males to uninfected females. As pointed out by both reviewers, this is a nice study, well written and easy to read. All the results are new and allow to better understand the role of the soft tick, Ornithodoros maritimus, as a potential vector of Babesia sp. YLG. They finally question about the degree to which the parasite can be maintained locally by ticks and the epidemiological consequences of infection for both O. maritimus and its avian host. For all these reasons, I chose to recommend this article for Peer Community In Infections. References
| A soft tick vector of *Babesia* sp. YLG in Yellow-legged gull (*Larus michahellis*) nests | Claire Bonsergent, Marion Vittecoq, Carole Leray, Maggy Jouglin, Marie Buysse, Karen D. McCoy, Laurence Malandrin | <p style="text-align: justify;"><em>Babesia </em>sp. YLG has recently been described in Yellow-legged gull (<em>Larus michahellis</em>) chicks and belongs to the Peircei clade in the new classification of Piroplasms. Here, we studied <em>Babesia <... |  | Ecology of hosts, infectious agents, or vectors, Eukaryotic pathogens/symbionts, Interactions between hosts and infectious agents/vectors, Parasites, Vectors | Thomas Pollet | 2023-03-29 14:33:40 | View |
MANAGING BOARD
Jorge Amich
Christine Chevillon
Fabrice Courtin
Christine Coustau
Thierry De Meeûs
Heather R. Jordan
Karl-Heinz Kogel
Yannick Moret
Thomas Pollet
Benjamin Roche
Benjamin Rosenthal
Bashir Salim
Lucy Weinert










