Latest recommendations

| Id | Title | Authors▼ | Abstract | Picture | Thematic fields | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
03 Nov 2023

Longitudinal Survey of Astrovirus infection in different bat species in Zimbabwe: Evidence of high genetic Astrovirus diversityVimbiso Chidoti, Helene De Nys, Malika Abdi, Getrudre Mashura, Valerie Pinarello, Ngoni Chiweshe, Gift Matope, Laure Guerrini, Davies Pfulenyi, Julien Cappelle, Ellen Mwandiringana, Dorothee Misse, Gori Elizabeth, Mathieu Bourgarel, Florian Liegeois https://doi.org/10.1101/2023.04.14.536987High diversity and evidence for inter-species transmission in astroviruses surveyed from bats in ZibabwaeRecommended by Tim James based on reviews by 2 anonymous reviewersMost infectious diseases of humans are zoonoses, and many of these come from particularly species diverse reservoir taxa, such as bats, birds, and rodents (1). Because of our changing landscape, there is increased exposure of humans to wildlife diseases reservoirs, yet we have little basic information about prevalence, hotspots, and transmission factors of most zoonotic pathogens. Viruses are particularly worrisome as a public health risk due to their fast mutation rates and well-known cross-species transmission abilities. There is a global push to better survey wildlife for viruses (2), but these studies are difficult, and the problem is vast. Astroviruses (AstVs) comprise a diverse family of ssRNA viruses known from mammals and birds. Astroviruses can cause gastroenteritis in humans and are more common in elderly and young children, but the relationship of human to non-human Astroviridae as well as transmission routes are unclear. AstVs have been detected at high prevalence in bats in multiple studies (3,4), but it is unclear what factors, such as co-infecting viruses and bat reproductive phenology, influence viral shedding and prevalence. References 1. Mollentze N, Streicker DG. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proceedings of the National Academy of Sciences. 2020 Apr 28;117(17):9423-30. https://doi.org/10.1073/pnas.1919176117 | Longitudinal Survey of Astrovirus infection in different bat species in Zimbabwe: Evidence of high genetic Astrovirus diversity | Vimbiso Chidoti, Helene De Nys, Malika Abdi, Getrudre Mashura, Valerie Pinarello, Ngoni Chiweshe, Gift Matope, Laure Guerrini, Davies Pfulenyi, Julien Cappelle, Ellen Mwandiringana, Dorothee Misse, Gori Elizabeth, Mathieu Bourgarel, Florian Liegeois | <p>Astroviruses (AstVs) have been discovered in over 80 animal species including diverse bat species and avian species. A study on Astrovirus circulation and diversity in different insectivorous and frugivorous chiropteran species roosting in tree... |  | Animal diseases, Epidemiology, Molecular genetics of hosts, infectious agents, or vectors, Reservoirs, Viruses, Zoonoses | Tim James | 2023-04-18 14:58:43 | View | |
14 Nov 2022
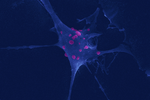
Ehrlichia ruminantium uses its transmembrane protein Ape to adhere to host bovine aortic endothelial cellsValérie Pinarello, Elena Bencurova, Isabel Marcelino, Olivier Gros, Carinne Puech, Mangesh Bhide, Nathalie Vachiery, Damien F. Meyer https://doi.org/10.1101/2021.06.15.447525Adhesion process of Ehrlichia ruminantium to its host cell: the role of the protein ERGACDS01230 elucidatedRecommended by Thomas Pollet based on reviews by Rodolfo García-Contreras and Alejandro Cabezas-CruzAs recently reported by the world organisation for animal health, 60% of infectious diseases are zoonotic with a significant part associated to ticks. Ticks can transmit various pathogens such as bacteria, viruses and parasites. Among pathogens known to be transmitted by ticks, Ehrlichia ruminantium is an obligate intracellular Gram-negative bacterium responsible for the fatal heartwater disease of domestic and wild ruminants (Allsopp, 2010). E. ruminantium is transmitted by ticks of the genus Amblyomma in the tropical and sub-Saharan areas, as well as in the Caribbean islands. It constitutes a major threat for the American livestock industries since a suitable tick vector is already present in the American mainland and potential introduction of infected A. variegatum through migratory birds or uncontrolled movement of animals from Caribbean could occur (i.e. Deem, 1998 ; Kasari et al 2010). The disease is also a major obstacle to the introduction of animals from heartwater-free to heartwater-infected areas into sub-Saharan Africa and thus restrains breeding programs aiming at upgrading local stocks (Allsopp, 2010). In this context, it is essential to develop control strategies against heartwater, as developing effective vaccines, for instance. Such an objective requires a better understanding of the early interaction of E. ruminantium and its host cells and of the mechanisms associated with bacterial adhesion to the host-cell. In this study, the authors. studied the role of E. ruminantium membrane protein ERGA_CDS_01230 in the adhesion process to host bovine aortic endothelial cells (BAEC). After successfully producing the recombinant version of the protein, Pinarello et al (2022) followed the in vitro culture of E. ruminantium in BAEC and observed that the expression of the protein peaked at the extracellular infectious elementary body stages. This result would suggest the likely involvement of the protein in the early interaction of E. ruminantium with its host cells. The authors then showed using flow cytometry, and scanning electron microscopy, that beads coated with the recombinant protein adhered to BAEC. In addition, they also observed that the adhesion protein of E. ruminantium interacted with proteins of the cell's lysate, membrane and organelle fractions. Additionally, enzymatic treatment, degrading dermatan and chondroitin sulfates on the surface of BAEC, was associated with a 50% reduction in the number of bacteria in the host cell after a development cycle, indicating that glycosaminoglycans might play a role in the adhesion of E. ruminantium to the host-cell. Finally, the authors observed that the adhesion protein of E. ruminantium induced a humoral response in vaccinated animals, making this protein a possible vaccine candidate. As rightly pointed out by both reviewers, the results of this study represent a significant advance (i) in the understanding of the role of the E. ruminantium membrane protein ERGA_CDS_01230 in the adhesion process to the host-cell and (ii) in the development of new control strategies against heartwater as this protein might potentially be used as an immunogen for the development of future vaccines. References Allsopp, B.A. (2010). Natural history of Ehrlichia ruminantium. Vet Parasitol 167, 123-135. https://doi.org/10.1016/j.vetpar.2009.09.014 Deem, S.L. (1998). A review of heartwater and the threat of introduction of Cowdria ruminantium and Amblyomma spp. ticks to the American mainland. J Zoo Wildl Med 29, 109-113. Kasari, T.R. et al (2010). Recognition of the threat of Ehrlichia ruminantium infection in domestic and wild ruminants in the continental United States. J Am Vet Med Assoc. 237:520-30. https://doi.org/10.2460/javma.237.5.520 Pinarello V, Bencurova E, Marcelino I, Gros O, Puech C, Bhide M, Vachiery N, Meyer DF (2022) Ehrlichia ruminantium uses its transmembrane protein Ape to adhere to host bovine aortic endothelial cells. bioRxiv, 2021.06.15.447525, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2021.06.15.447525 | *Ehrlichia ruminantium* uses its transmembrane protein Ape to adhere to host bovine aortic endothelial cells | Valérie Pinarello, Elena Bencurova, Isabel Marcelino, Olivier Gros, Carinne Puech, Mangesh Bhide, Nathalie Vachiery, Damien F. Meyer | <p><em>Ehrlichia ruminantium</em> is an obligate intracellular bacterium, transmitted by ticks of the genus <em>Amblyomma</em> and responsible for heartwater, a disease of domestic and wild ruminants. High genetic diversity of <em>E. ruminantium</... |  | Interactions between hosts and infectious agents/vectors, Microbiology of infections | Thomas Pollet | Rodolfo García-Contreras, Alejandro Cabezas-Cruz | 2021-10-14 16:54:54 | View |
28 Sep 2023
Influence of endosymbionts on the reproductive fitness of the tick Ornithodoros moubataTaraveau Florian, Pollet Thomas, Duhayon Maxime, Gardès Laëtitia, Jourdan-Pineau Hélène https://doi.org/10.1101/2023.05.09.539061The cost of endosymbionts on the reproductive fitness of the soft tick Ornithodoros moubataRecommended by Angélique Gobet based on reviews by Luciana Raggi Hoyos and Tuomas Aivelo based on reviews by Luciana Raggi Hoyos and Tuomas Aivelo
Ticks are amongst the most important pathogen vectors in medical and veterinary clinical settings worldwide (Dantas-Torres et al., 2012). Like other holobionts, ticks live in association with a diverse microbiota. It includes tick-borne pathogens (TBP) and other microorganisms that have a beneficial or detrimental effect on the physiology of the host and can also affect the transmission of TBP to animals or humans. In this microbiota, primary endosymbionts, which are obligatory and inheritable, play a role in tick reproduction, the host defense and adaptation to varying environmental conditions (Duron et al., 2018). However, the effect of the microbiota structure and of the endosymbionts on tick fitness and reproduction is not well known. The soft tick Ornithodoros moubata, a parasite known to transmit African swine fever virus (Vial, 2009), is known to host Francisella-like and Rickettsia endosymbionts (Duron et al., 2018). These endosymbionts carry genes involved in B vitamin synthesis which may be supplemented to the host (Bonnet & Pollet, 2021). Here, the authors investigated the role of endosymbionts on the reproductive fitness of Ornithodoros moubata by conducting two experiments (Taraveau et al., 2023). First, they tested the effect of antibiotic treatment of 366 first-stage nymphs on the main endosymbionts Francisella-like and Rickettsia, and measured the endosymbionts presence overtime by qPCR. Second, they surveyed the effect of antibiotic treatment with or without the addition of B vitamins on the survival and reproductive fitness of 132 females over 50 days. This second experiment intended to identify whether the endosymbionts have an effect on the host reproduction or on its nutrition. The supplementation of B vitamin did not have a drastic effect on tick fitness or reproductive traits. However, antibiotic treatments reduced the presence of endosymbionts while increasing tick survival, suggesting a potential cost of hosting endosymbionts on the tick fitness. The authors did a lot of work to thoroughly follow the propositions from Dr Raggi, Dr Aivelo and myself to reconstruct and to revise the manuscript. I believe that the manuscript now reads very well and the answers to the reviews also add some value to the manuscript. As Dr Aivelo pointed out, “this study follows the traditional path of so-called population perturbation studies, where ecologists have administered antibiotics or antihelminths to different animals and seen how the community changes and what effects this has on the host fitness and survival”. As both reviewers stated, results from this study are valuable and provide important basic knowledge that will likely help conduct future experiments on tick microbiota. This recommendation is the result of the thorough reviewing work of Dr Aivelo and Dr Raggi which I warmly thank. Bonnet, S. I., & Pollet, T. (2021). Update on the intricate tango between tick microbiomes and tick‐borne pathogens. Parasite Immunology, 43(5), e12813. https://doi.org/10.1111/pim.12813 Dantas-Torres, F., Chomel, B. B., & Otranto, D. (2012). Ticks and tick-borne diseases: A One Health perspective. Trends in Parasitology, 28(10), 437–446. https://doi.org/10.1016/j.pt.2012.07.003 Duron, O., Morel, O., Noël, V., Buysse, M., Binetruy, F., Lancelot, R., Loire, E., Ménard, C., Bouchez, O., Vavre, F., & Vial, L. (2018). Tick-Bacteria Mutualism Depends on B Vitamin Synthesis Pathways. Current Biology, 28(12), 1896-1902.e5. https://doi.org/10.1016/j.cub.2018.04.038 Taraveau, F., Pollet, T., Duhayon, M., Gardès, L., & Jourdan-Pineau, H. (2023). Influence of endosymbionts on the reproductive fitness of the tick Ornithodoros moubata. bioRxiv, ver.3, peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2023.05.09.539061 Vial, L. (2009). Biological and ecological characteristics of soft ticks (Ixodida: Argasidae) and their impact for predicting tick and associated disease distribution. Parasite, 16(3), 191–202. https://doi.org/10.1051/parasite/2009163191 | Influence of endosymbionts on the reproductive fitness of the tick *Ornithodoros moubata* | Taraveau Florian, Pollet Thomas, Duhayon Maxime, Gardès Laëtitia, Jourdan-Pineau Hélène | <p style="text-align: justify;">Over the past decade, many studies have demonstrated the crucial role of the tick microbiome in tick biology. The soft tick <em>Ornithodoros moubata</em> is a hematophagous ectoparasite of <em>Suidae</em>, best know... | Mutualistic symbionts, Parasites, Pathogenic/Symbiotic Bacteria, Physiology of hosts, infectious agents, or vectors, Vectors | Angélique Gobet | 2023-05-25 19:00:33 | View | ||
07 Oct 2022

Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pestsS. Massart, I. Adams, M. Al Rwahnih, S. Baeyen, G. J. Bilodeau, A. G. Blouin, N. Boonham, T. Candresse, A. Chandelier, K. De Jonghe, A. Fox, Y.Z.A. Gaafar, P. Gentit, A. Haegeman, W. Ho, O. Hurtado-Gonzales, W. Jonkers, J. Kreuze, D. Kutjnak, B. B. Landa, M. Liu, F. Maclot, M. Malapi-Wight, H. J. Maree, F. Martoni, N. Mehle, A. Minafra, D. Mollov, A. G. Moreira, M. Nakhla, F. Petter, A.M. Piper, J. P. Ponchart, R. Rae, B. Remenant, Y. Rivera, B. Rodoni, M. Botermans, J.W. Roenhorst, J. Rollin , ... https://doi.org/10.5281/zenodo.7142136High-throughput sequencing for the diagnostic of plant pathologies and identification of pests: recommendations and challengesRecommended by Olivier Schumpp based on reviews by Denise Altenbach and David RoquisHigh-throughput sequencing (HTS) has revealed an incredible diversity of microorganisms in ecosystems and is also changing the monitoring of macroorganism biodiversity (Deiner et al. 2017; Piper et al. 2019). The diagnostic of plant pathogens and the identification of pests is gradually integrating the use of these techniques, but there are still obstacles. Most of them are related to the reliability of these analyses, which have long been considered insufficient because of their dependence on a succession of sophisticated operations involving parameters that are sometimes difficult to adapt to complex matrices or certain diagnostic contexts. The need to validate HTS approaches is gradually being highlighted in recent work but remains poorly documented (Bester et al. 2022). In this paper, a large community of experts presents and discusses the key steps for optimal control of HTS performance and reliability in a diagnostic context (Massart et al. 2022). It also addresses the issue of costs. The article provides recommendations that closely combine the quality control requirements commonly used in conventional diagnostics with newer or HTS-specific control elements and concepts that are not yet widely used. It discusses the value of these for the use of the various techniques currently covered by the terms "High Throughput Sequencing" in diagnostic activities. The elements presented are intended to limit false positive or false negative results but will also optimise the interpretation of contentious results close to the limits of analytical sensitivity or unexpected results, both of which appear to be frequent when using HTS. Furthermore, the need for risk analysis, verification and validation of methods is well illustrated with numerous examples for each of the steps considered crucial to ensure reliable use of HTS. The clear contextualisation of the proposals made by the authors complements and clarifies the need for user expertise according to the experimental objectives. Some unanswered questions that will require further development and validation are also presented. This article should benefit a large audience including researchers with some level of expertise in HTS but unfamiliar with the recent concepts of controls common in the diagnostic world as well as scientists with strong diagnostic expertise but less at ease with the numerous and complex procedures associated with HTS. References Bester R, Steyn C, Breytenbach JHJ, de Bruyn R, Cook G, Maree HJ (2022) Reproducibility and Sensitivity of High-Throughput Sequencing (HTS)-Based Detection of Citrus Tristeza Virus and Three Citrus Viroids. Plants, 11, 1939. https://doi.org/10.3390/plants11151939 Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26, 5872–5895. https://doi.org/10.1111/mec.14350 Massart, S et al. (2022) Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests. Zenodo, 6637519, ver. 3 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.6637519 Piper AM, Batovska J, Cogan NOI, Weiss J, Cunningham JP, Rodoni BC, Blacket MJ (2019) Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience, 8, giz092. https://doi.org/10.1093/gigascience/giz092 | Guidelines for the reliable use of high throughput sequencing technologies to detect plant pathogens and pests | S. Massart, I. Adams, M. Al Rwahnih, S. Baeyen, G. J. Bilodeau, A. G. Blouin, N. Boonham, T. Candresse, A. Chandelier, K. De Jonghe, A. Fox, Y.Z.A. Gaafar, P. Gentit, A. Haegeman, W. Ho, O. Hurtado-Gonzales, W. Jonkers, J. Kreuze, D. Kutjnak, B. B... | <p style="text-align: justify;">High-throughput sequencing (HTS) technologies have the potential to become one of the most significant advances in molecular diagnostics. Their use by researchers to detect and characterize plant pathogens and pests... |  | Diagnosis, Pest management, Phytopathology, Plant diseases | Olivier Schumpp | 2022-06-13 11:26:18 | View | |
29 Jan 2024
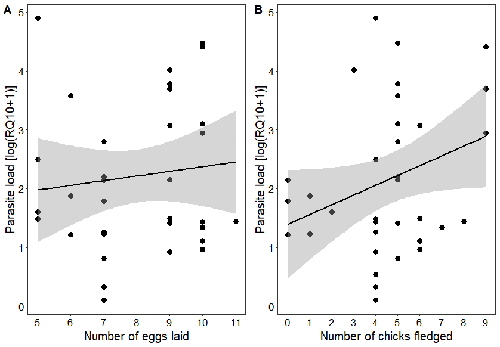
Spring reproductive success influences autumnal malarial load in a passerine birdRomain Pigeault, Camille-Sophie Cozzarolo, Jérôme Wassef, Jérémy Gremion, Marc Bastardot, Olivier Glaizot, Philippe Christe https://doi.org/10.1101/2023.07.28.550923Avian Plasmodium parasitaemia as an indicator of reproduction investmentRecommended by Claire Loiseau based on reviews by Luz García-Longoria and 2 anonymous reviewers based on reviews by Luz García-Longoria and 2 anonymous reviewers
Effects of the seasonal variations on within-host parasitaemia are still not well understood and potentially due to numerous factors, e.g. host and parasite species, host sex or age, or geographical regions. In this study, over three years in Switzerland, Pigeault et al. (2024) collected data on great tits reproductive outputs – laying date, clutch size, fledging success – to determine whether they were associated with avian Plasmodium parasitaemia before (winter), during (spring) and after (autumn) the breeding season. They focused on two lineages from two species: a highly generalist lineage Plasmodium relictum (lineage SGS1; Bensch et al. 2009) and a more specialized lineage Plasmodium homonucleophilum (lineage SW2). As previously found, they showed that parasitaemia level is low during the winter and then increase in spring (Applegate, 1970; Applegate 1971). Spring recurrences have been intensively studied but are still not well understood since many non-exclusive factors can provoke them, i.e environmental stressors, reproductive hormones, co-infections or bites of mosquitoes (Cornet et al. 2014). Interestingly, the parasitaemia level during the winter before and during the breeding season were not associated to the reproductive success, meaning that birds in their populations with low parasitaemia during the winter had not more fledglings than the ones with a higher parasitaemia. However, the individuals who invested the most in the reproduction with a higher number of fledglings had also a higher parasitaemia in the following autumn. The number of laid eggs was not associated with the parasitaemia during the following autumn, showing that the initial investment in the reproduction is less important than the parental care (e.g. chicks feeding) in terms of mid/long term cost. The originality here is that authors followed populations during three periods of the year, which is not an easy task and rarely done in natural populations. Their results highlight the mid/long-term effect of higher resource allocation into reproduction on individuals’ immune system and ability to control parasite replication. Further analyses on various lineages and bird populations from other geographical regions (i.e. different latitudes) would be the next relevant step. References Applegate JE (1971) Spring relapse of Plasmodium relictum infections in an experimental field population of English sparrows (Passer domesticus). Journal of Wildlife Diseases, 7, 37–42. https://doi.org/10.7589/0090-3558-7.1.37 Applegate JE, Beaudoin RL (1970) Mechanism of spring relapse in avian malaria: Effect of gonadotropin and corticosterone. Journal of Wildlife Diseases, 6, 443–447. https://doi.org/10.7589/0090-3558-6.4.443 Bensch S, Hellgren O, Pérez‐Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Molecular Ecology Resources, 9, 1353-1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x Cornet S, Nicot A, Rivero A, Gandon S (2014) Evolution of plastic transmission strategies in avian malaria. PLoS Pathogens, 10, e1004308. https://doi.org/10.1371/journal.ppat.1004308 Pigeault R, Cozzarolo CS, Wassef J, Gremion J, Bastardot M, Glaizot O, Christe P (2024) Spring reproductive success influences autumnal malarial load in a passerine bird. bioRxiv ver 3. Peer reviewed and recommended by Peer Community In Infections. https://doi.org/10.1101/2023.07.28.550923 | Spring reproductive success influences autumnal malarial load in a passerine bird | Romain Pigeault, Camille-Sophie Cozzarolo, Jérôme Wassef, Jérémy Gremion, Marc Bastardot, Olivier Glaizot, Philippe Christe | <p>Although avian haemosporidian parasites are widely used as model organisms to study fundamental questions in evolutionary and behavorial ecology of host-parasite interactions, some of their basic characteristics, such as seasonal variations in ... |  | Interactions between hosts and infectious agents/vectors, Parasites | Claire Loiseau | Carolina Chagas, Anonymous, Luz García-Longoria | 2023-08-11 14:14:56 | View |
29 Jan 2024
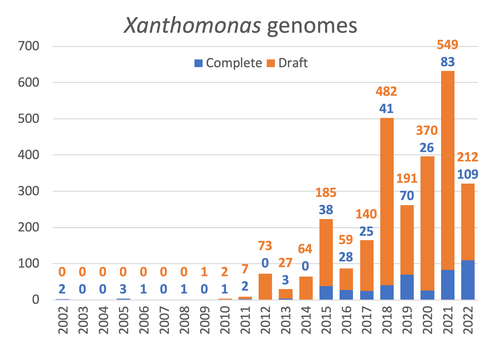
Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy, Provided Insight into the Emergence of Pathogenic Bacteria, Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on XanthomonadsRalf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek https://doi.org/10.5281/zenodo.8223857Advancing Pathogen Genomics: A Comprehensive Review of the Xanthomonas(*) Genome's Impact on Bacterial Research and Control StrategiesRecommended by Damien François Meyer based on reviews by Boris Vinatzer and 3 anonymous reviewers based on reviews by Boris Vinatzer and 3 anonymous reviewers
The paper titled "Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy Provided Insight into the Emergence of Pathogenic Bacteria Enabled New Fundamental Discoveries and Helped Developing Novel Control Measures – A Perspective from the French Network on Xanthomonads" by Ralf Koebnik et al. (2023) is an insightful contribution to the field of genomics and its application in understanding pathogenic bacteria, particularly Xanthomonas. This comprehensive review offers a unique perspective from the French Network on Xanthomonads, underscoring the significant advancements in taxonomy, pathogen emergence, and development of control strategies due to genomic research. One of the paper's main strengths is its thorough exploration of how genomics has revolutionized our understanding of Xanthomonas and other pathogenic bacteria. It sheds light on the evolution and emergence of these pathogens, contributing significantly to the development of novel and effective control measures. The authors' detailed account of the historical progress and current state of genomics in this field highlights its pivotal role in guiding future research and practical applications in managing bacterial diseases. Moreover, the paper emphasizes the importance of collaborative efforts and the sharing of knowledge within scientific networks, as exemplified by the French Network on Xanthomonas. This approach not only enriches the study but also serves as a model for future collaborative research endeavors. In conclusion, the work of Koebnik et al. is a valuable resource for researchers, policymakers, and practitioners in the field of plant pathology and genomics. It not only provides a comprehensive overview of the advances in genomics related to Xanthomonas but also illustrates the broader impact of genomic studies in understanding and managing pathogenic bacteria. References Ralf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek (2024) Celebrating the 20th anniversary of the first Xanthomonas genome gequences – How genomics revolutionized taxonomy, provided insight into the emergence of pathogenic bacteria, enabled new fundamental discoveries and helped developing novel control measures – A perspective from the French network on Xanthomonads. Zenodo ver. 3, peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.5281/zenodo.8223857 | Celebrating the 20th Anniversary of the First Xanthomonas Genome Sequences – How Genomics Revolutionized Taxonomy, Provided Insight into the Emergence of Pathogenic Bacteria, Enabled New Fundamental Discoveries and Helped Developing Novel Control ... | Ralf Koebnik, Sophie Cesbron, Nicolas W. G. Chen, Marion Fischer-Le Saux, Mathilde Hutin, Marie-Agnès Jacques, Laurent D. Noël, Alvaro Perez-Quintero, Perrine Portier, Olivier Pruvost, Adrien Rieux, And Boris Szurek | <p>In this Opinion paper, members of the French Network on Xanthomonads give their personal view on what they consider to be some of the groundbreaking discoveries in the field of molecular plant pathology over the past 20 years. By celebrating th... |  | Epidemiology, Evolution of hosts, infectious agents, or vectors, Genomics, functional genomics of hosts, infectious agents, or vectors, Interactions between hosts and infectious agents/vectors, Molecular biology of infections, Molecular genetics o... | Damien François Meyer | 2023-08-09 10:37:15 | View | |
14 Dec 2022

Transcriptome responses of the aphid vector Myzus persicae are shaped by identities of the host plant and the virusQuentin Chesnais, Victor Golyaev, Amandine Velt, Camille Rustenholz, Maxime Verdier, Véronique Brault, Mikhail M. Pooggin, Martin Drucker https://doi.org/10.1101/2022.07.18.500449How do multiple host plants and virus species challenge aphid molecular machinery?Recommended by Sebastien Massart based on reviews by Juan José Lopez Moya and Michelle HeckThe impact of virus infection of a plant on an aphid’s behaviour has been observed in many studies [1]. Indeed, virus infection can alter plant biochemistry through the emission of volatile organic compounds and plant tissue content modification. These alterations can further impact the interactions between plants and aphids. However, although it is a well-known phenomenon, very few studies have explored the consequences of plant virus infection on the gene expression of aphids to understand better the aphid’s manipulation by the plant virus. In this context, the recommended study [2] reports a comprehensive transcriptomic analysis of the genes expressed by one aphid species, Myzus persicae, a vector of several plant viruses, when feeding on plants. Michelle Heck underlined how significant this study is for comprehending the molecular bases of aphid-vector manipulation by plant viruses (see below). Interestingly, the study design has integrated several factors that might influence the gene expression of M. persicae when feeding on the plant. Indeed, the authors investigated the effect of two plant species (Arabidopsis thaliana and Camelia sativa) and two virus species [turnip yellows virus (TuYV) and cauliflower mosaic virus (CaMV)]. Noteworthy, the transmission mode of TuYV is circulative and persistent, while CaMV is transmitted by a semi-persistent non-circulative mode. As Juan José Lopez Moya mentioned, multiple comparisons allowed the identification of the different responses of aphids in front of different host plants infected or not by different viruses (see below). This publication is complementary to a previous publication from the same team focusing on plant transcriptome analysis [3]. Thanks to their experimental design, the authors identified genes commonly deregulated by both viruses and/or both plant species and deregulated genes by a single virus or a single plant. Figure 4 nicely summarizes the number of deregulated genes. A thorough discussion on the putative role of deregulated genes in different conditions gave a comprehensive follow-up of the results and their impact on the current knowledge of plant-virus-vector interactions. This study has now opened the gate to promising research focusing on the functional validation of the identified genes while also narrowing the study from the body to the tissue level. References: 1. Carr JP, Tungadi T, Donnelly R, Bravo-Cazar A, Rhee S-J, Watt LG, Mutuku JM, Wamonje FO, Murphy AM, Arinaitwe W, Pate AE, Cunniffe NJ, Gilligan CA (2020) Modelling and manipulation of aphid-mediated spread of non-persistently transmitted viruses. Virus Research, 277, 197845. https://doi.org/10.1016/j.virusres.2019.197845 2. Chesnais Q, Golyaev V, Velt A, Rustenholz C, Verdier M, Brault V, Pooggin MM, Drucker M (2022) Transcriptome responses of the aphid vector Myzus persicae are shaped by identities of the host plant and the virus. bioRxiv , 2022.07.18.500449, ver. 5 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.07.18.500449 3. Chesnais Q, Golyaev V, Velt A, Rustenholz C, Brault V, Pooggin MM, Drucker M (2022) Comparative Plant Transcriptome Profiling of Arabidopsis thaliana Col-0 and Camelina sativa var. Celine Infested with Myzus persicae Aphids Acquiring Circulative and Noncirculative Viruses Reveals Virus- and Plant-Specific Alterations Relevant to Aphid Feeding Behavior and Transmission. Microbiology Spectrum, 10, e00136-22. https://doi.org/10.1128/spectrum.00136-22 | Transcriptome responses of the aphid vector *Myzus persicae* are shaped by identities of the host plant and the virus | Quentin Chesnais, Victor Golyaev, Amandine Velt, Camille Rustenholz, Maxime Verdier, Véronique Brault, Mikhail M. Pooggin, Martin Drucker | <p style="text-align: justify;"><strong>Background:</strong> Numerous studies have documented modifications in vector orientation behavior, settling and feeding behavior, and/or fecundity and survival due to virus infection in host plants. These a... |  | Behaviour of hosts, infectious agents, or vectors, Cell biology of hosts, infectious agents, or vectors, Molecular biology of infections, Physiology of hosts, infectious agents, or vectors, Phytopathology, Plant diseases, Vectors, Viruses | Sebastien Massart | 2022-07-19 15:24:14 | View | |
24 Jan 2024
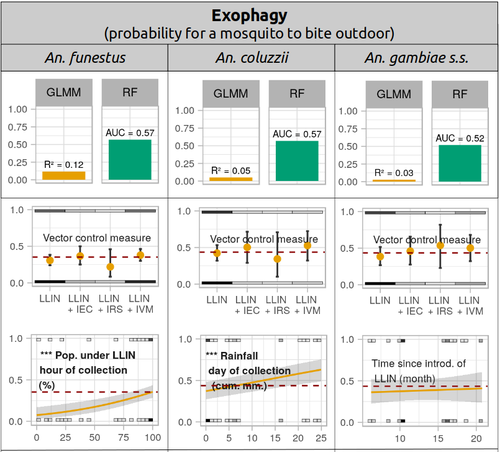
Physiological and behavioural resistance of malaria vectors in rural West-Africa : a data mining study to address their fine-scale spatiotemporal heterogeneity, drivers, and predictabilityPaul Taconet, Dieudonné Diloma Soma, Barnabas Zogo, Karine Mouline, Frédéric Simard, Alphonsine Amanan Koffi, Roch Kounbobr Dabiré, Cédric Pennetier, Nicolas Moiroux https://doi.org/10.1101/2022.08.20.504631Large and complete datasets, and modelling reveal the major determinants of physiological and behavioral insecticide resistance of malaria vectorsRecommended by Thierry DE MEEÛS based on reviews by Haoues Alout and 1 anonymous reviewer based on reviews by Haoues Alout and 1 anonymous reviewer
Parasites represent the most diverse and adaptable ecological group of the biosphere (Timm & Clauson, 1988; De Meeûs et al., 1998; Poulin & Morand, 2000; De Meeûs & Renaud, 2002). The human species is known to considerably alter biodiversity, though it hosts, and thus sustains the maintenance of a spectacular diversity of parasites (179 species for eukaryotic species only) (De Meeûs et al., 2009). Among these, the five species of malaria agents (genus Plasmodium) remain a major public health issue around the world. Plasmodium falciparum is the most prevalent and lethal of these (Liu et al., 2010). With a pick of up to 2 million deaths due to malaria in 2004, deaths decreased to around 1 million in 2010 (Murray et al., 2012), to reach 619,000 in 2021, most of which in sub-Saharan Africa, and 79% of which were among children aged under 5 years (World Health Organization, 2022). As stressed by Taconet et al. (2023), reduction in malaria deaths is attributable to control measures, in particular against its vectors (mosquitoes of the genus Anopheles). Nevertheless, the success of vector control is hampered by several factors (biological, environmental and socio-economic), and in particular by the great propensity of targeted mosquitoes to evolve physiological or behavioral avoidance of anti-vectorial measures. In their paper Taconet et al. (2023) aims at understanding what are the main factors that determine the evolution of insecticide resistance in several malaria vectors, in relation to the biological determinisms of behavioral resistance and how fast such evolutions take place. To tackle these objectives, authors collected an impressive amount of data in two rural areas of West Africa. With appropriate modeling, Taconet et al. discovered, among many other results, a predominant role of public health measures, as compared to agricultural practices, in the evolution of physiological resistance. They also found that mosquito foraging activities are mostly explained by host availability and climate, with a poor, if any, association with genetic markers of physiological resistance to insecticides. These findings represent an important contribution to the field and should help at designing more efficient control strategies against malaria.
References De Meeûs T, Michalakis Y, Renaud F (1998) Santa Rosalia revisited: or why are there so many kinds of parasites in “the garden of earthly delights”? Parasitology Today, 14, 10–13. https://doi.org/10.1016/S0169-4758(97)01163-0 De Meeûs T, Prugnolle F, Agnew P (2009) Asexual reproduction in infectious diseases. In: Lost Sex: The Evolutionary Biology of Parthenogenesis (eds Schön I, Martens K, van Dijk P), pp. 517-533. Springer, NY. https://doi.org/10.1007/978-90-481-2770-2_24 De Meeûs T, Renaud F (2002) Parasites within the new phylogeny of eukaryotes. Trends in Parasitology, 18, 247–251. https://doi.org/10.1016/S1471-4922(02)02269-9 Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, Ndjango JB, Sanz CM, Morgan DB, Locatelli S, Gonder MK, Kranzusch PJ, Walsh PD, Delaporte E, Mpoudi-Ngole E, Georgiev AV, Muller MN, Shaw GM, Peeters M, Sharp PM, Rayner JC, Hahn BH (2010) Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature, 467, 420–425. https://doi.org/10.1038/nature09442 Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, Fullman N, Naghavi M, Lozano R, Lopez AD (2012) Global malaria mortality between 1980 and 2010: a systematic analysis. The Lancet, 379, 413–431. https://doi.org/10.1016/S0140-6736(12)60034-8 Poulin R, Morand S (2000) The diversity of parasites. Quarterly Review of Biology, 75, 277–293. https://doi.org/10.1086/393500 Taconet P, Soma DD, Zogo B, Mouline K, Simard F, Koffi AA, Dabire RK, Pennetier C, Moiroux N (2023) Physiological and behavioural resistance of malaria vectors in rural West-Africa : a data mining study to address their fine-scale spatiotemporal heterogeneity, drivers, and predictability. bioRxiv, ver. 4 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.08.20.504631 Timm RM, Clauson BL (1988) Coevolution: Mammalia. In: 1988 McGraw-Hill yearbook of science & technology, pp. 212–214. McGraw-Hill Book Company, New York. World Health Organization (2022) World malaria report 2022. Geneva: World Health Organization; 2022. Licence: CC BY-NC-SA 3.0 IGO. https://iris.who.int/bitstream/handle/10665/365169/9789240064898-eng.pdf?sequence=1.
| Physiological and behavioural resistance of malaria vectors in rural West-Africa : a data mining study to address their fine-scale spatiotemporal heterogeneity, drivers, and predictability | Paul Taconet, Dieudonné Diloma Soma, Barnabas Zogo, Karine Mouline, Frédéric Simard, Alphonsine Amanan Koffi, Roch Kounbobr Dabiré, Cédric Pennetier, Nicolas Moiroux | <p>Insecticide resistance and behavioural adaptation of malaria mosquitoes affect the efficacy of long-lasting insecticide nets - currently the main tool for malaria vector control. To develop and deploy complementary, efficient and cost-effective... |  | Behaviour of hosts, infectious agents, or vectors, Ecology of hosts, infectious agents, or vectors, Pesticide resistance, Population genetics of hosts, infectious agents, or vectors, Vectors | Thierry DE MEEÛS | Haoues Alout, Anonymous | 2023-07-03 11:29:10 | View |
21 Jul 2022

Structural variation turnovers and defective genomes: key drivers for the in vitro evolution of the large double-stranded DNA koi herpesvirus (KHV)Nurul Novelia Fuandila, Anne-Sophie Gosselin-Grenet, Marie-Ka Tilak, Sven M Bergmann, Jean-Michel Escoubas, Sandro Klafack, Angela Mariana Lusiastuti, Munti Yuhana, Anna-Sophie Fiston-Lavier, Jean-Christophe Avarre, Emira Cherif https://doi.org/10.1101/2022.03.10.483410Understanding the in vitro evolution of Cyprinid herpesvirus 3 (CyHV-3), a story of structural variations that can lead to the design of attenuated virus vaccinesRecommended by Jorge Amich based on reviews by Lucie Cappuccio and Veronique Hourdel based on reviews by Lucie Cappuccio and Veronique Hourdel
Structural variations (SVs) play a key role in viral evolution, and therefore they are also important for infection dynamics. However, the contribution of structural variations to the evolution of double-stranded viruses is limited. This knowledge can help to understand the population dynamics and might be crucial for the future development of viral attenuated vaccines. In this study, Fuandila et al (1) use the Cyprinid herpesvirus 3 (CyHV-3), commonly known as koi herpesvirus (KHV), to investigate the variability and contribution of structural variations (SV) for viral evolution after 99 passages in vitro. This virus, with the largest genome among herperviruses, causes a lethal infection in common carp and koi associated with mortalities up to 95% (2). Interestingly, KHV infections are caused by haplotype mixtures, which possibly are a source of genome diversification, but make genomic comparisons more difficult. The authors have used ultra-deep long-read sequencing of two passages, P78 and P99, which were previously described to have differences in virulence. They have found a surprisingly high and wide distribution of SVs along the genome, which were enriched in inversion and deletion events and that often led to defective viral genomes. Although it is known that these defective viral genomes negatively impact viral replication, their implications for virus persistence are still unclear. Subsequently, the authors concentrated on the virulence-relevant region ORF150, which was found to be different in P78 (deletion in 100% of the reads) and P99 (reference-like haplotype). To understand this loss and gain of full ORF150, they searched for SV turn-over in 10 intermediate passages. This analysis revealed that by passage 10 deleted and inverted (attenuated) haplotypes had already appeared, steadily increased frequency until P78, and then completely disappeared between P78 and P99. This is a striking result that raises new questions as to how this clearance occurs, which is really important as these reversions may result in undesirable increases in virulence of live-attenuated vaccines. We recommend this preprint because its use of ultra-deep long-read sequencing has permitted to better understand the role of SV diversity and dynamics in viral evolution. This study shows an unexpectedly high number of structural variations, revealing a novel source of virus diversification and confirming the different mixtures of haplotypes in different passages, including the gain of function. This research provides basic knowledge for the future design of live-attenuated vaccines, to prevent the reversion to virulent viruses. References (1) Fuandila NN, Gosselin-Grenet A-S, Tilak M-K, Bergmann SM, Escoubas J-M, Klafack S, Lusiastuti AM, Yuhana M, Fiston-Lavier A-S, Avarre J-C, Cherif E (2022) Structural variation turnovers and defective genomes: key drivers for the in vitro evolution of the large double-stranded DNA koi herpesvirus (KHV). bioRxiv, 2022.03.10.483410, ver. 4 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2022.03.10.483410 (2) Sunarto A, McColl KA, Crane MStJ, Sumiati T, Hyatt AD, Barnes AC, Walker PJ. Isolation and characterization of koi herpesvirus (KHV) from Indonesia: identification of a new genetic lineage. Journal of Fish Diseases, 34, 87-101. https://doi.org/10.1111/j.1365-2761.2010.01216.x | Structural variation turnovers and defective genomes: key drivers for the in vitro evolution of the large double-stranded DNA koi herpesvirus (KHV) | Nurul Novelia Fuandila, Anne-Sophie Gosselin-Grenet, Marie-Ka Tilak, Sven M Bergmann, Jean-Michel Escoubas, Sandro Klafack, Angela Mariana Lusiastuti, Munti Yuhana, Anna-Sophie Fiston-Lavier, Jean-Christophe Avarre, Emira Cherif | <p style="text-align: justify;">Structural variations (SVs) constitute a significant source of genetic variability in virus genomes. Yet knowledge about SV variability and contribution to the evolutionary process in large double-stranded (ds)DNA v... |  | Animal diseases, Evolution of hosts, infectious agents, or vectors, Genomics, functional genomics of hosts, infectious agents, or vectors, Viruses | Jorge Amich | Lucie Cappuccio, Veronique Hourdel | 2022-03-11 10:50:50 | View |
19 Feb 2024
Population genetics of Glossina palpalis gambiensis in the sleeping sickness focus of Boffa (Guinea) before and after eight years of vector control: no effect of control despite a significant decrease of human exposure to the diseaseMoise S. Kagbadouno, Modou Séré, Adeline Ségard, Abdoulaye Dansy Camara, Mamadou Camara, Bruno Bucheton, Jean-Mathieu Bart, Fabrice Courtin, Thierry de Meeûs, Sophie Ravel https://doi.org/10.1101/2023.07.25.550445Reaching the last miles for transmission interruption of sleeping sickness in Guinea: follow-up of achievements and policy making using microsatellites-based population geneticsRecommended by Hugues Nana Djeunga based on reviews by Fabien HALKETT and 2 anonymous reviewers based on reviews by Fabien HALKETT and 2 anonymous reviewers
Thanks to the coordinated and sustained efforts of national control programs, the World Health Organization (WHO), bilateral cooperation and nongovernmental organizations, the incidence of Human African Trypanosomiasis (HAT), better known as sleeping sickness, has drastically decreased during the last two decades (WHO, 2023a). Indeed, between 1999 and 2022, the reported number of new cases of the chronic form of sleeping sickness (Trypanosoma brucei gambiense) fell by 97% (from 27 862 to 799), and the number of newly reported cases of the acute form of HAT (Trypanosoma brucei rhodesiense) fell by 94% (from 619 to 38) (WHO, 2023b). These encouraging trends led the WHO to target this debilitating and highly fatal (if untreated) vector-borne parasitic disease for elimination as a public health problem by 2020, and for interruption of transmission (zero case) by 2030 (WHO, 2021, WHO, 2023a). However, the disease is persisting in many foci, and even some cases of resurgence have been documented after unfortunate events such as war or pandemics (Moore et al., 1999; Sah et al., 2023. Simarro et al). Although effective control measures, diagnosis and treatment are complex and require specific skills (WHO, 2023), especially in a context which animal reservoirs, including hidden reservoirs, can contribute to the maintenance/persistence of infection (Welburn and Maudlin, 2012; Camara et al., 2021). Vector control therefore appears as a viable alternative to accelerate sleeping sickness transmission interruption, and WHO has identified some critical actions for HAT elimination, including the coordination of vector control and animal trypanosomiasis management among countries, stakeholders and other sectors (e.g. tourism and wildlife) through multisectoral national bodies to maximize synergies (WHO, 2021). The paper by Kagbadouno and Collaborators (2024) uses microsatellite markers genotyping and population genetics tools to investigate the impact of 11 years of tiny target-based vector control on the population biology of Glossina palpalis gambiensis in Boffa, one of the three active sleeping sickness foci in Guinea (Kagbadouno et al., 2012). Although vector control significantly reduced the apparent densities of tsetse flies (and therefore the human exposure to the vector) as well as the prevalence and incidence of the disease in the Boffa HAT focus (Courtin et al., 2015), no genetic signature of vector control was observed as no difference in population size, before and after the onset of the control policy, was found. The authors then provided national programs and implementing partners with indications on the actions to be taken to (i) maintain the achievements of vector control (thus avoiding rebound/resurgence as was experienced in the past (Franco et al., 2014), and (ii) accelerate the momentum towards elimination by for example combining these vector control efforts with medical surveys for case detection and treatment, in line with WHO recommendations (WHO, 2021). References Camara M, Soumah AM, Ilboudo H, Travaillé C, Clucas C, Cooper A, Kuispond Swar NR, Camara O, Sadissou I, Calvo Alvarez E, Crouzols A, Bart JM, Jamonneau V, Camara M, MacLeod A, Bucheton B, Rotureau B. Extravascular Dermal Trypanosomes in Suspected and Confirmed Cases of gambiense Human African Trypanosomiasis. Clin Infect Dis. 2021 Jul 1;73(1):12-20. https://doi.org/10.1093/cid/ciaa897 Courtin F, Camara M, Rayaisse JB, Kagbadouno M, Dama E, Camara O, Traore IS, Rouamba J, Peylhard M, Somda MB, Leno M, Lehane MJ, Torr SJ, Solano P, Jamonneau V, Bucheton B (2015) Reducing human-tsetse contact significantly enhances the efficacy of sleeping sickness active screening campaigns: a promising result in the context of elimination. PLoS Neglected Tropical Diseases, 9. https://doi.org/10.1371/journal.pntd.0003727 Franco JR, Simarro PP, Diarra A, Jannin JG. (2014) Epidemiology of human African trypanosomiasis. Clin Epidemiol. 6:257-75. https://doi.org/10.2147/CLEP.S39728 Kagbadouno, M. S., Séré, M., Ségard, A., Camara, A. D., Camara, M., Bucheton, B., ... & Ravel, S. (2023). Population genetics of Glossina palpalis gambiensis in the sleeping sickness focus of Boffa (Guinea) before and after eight years of vector control: no effect of control despite a significant decrease of human exposure to the disease. bioRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Infections. https://doi.org/10.1101/2023.07.25.550445 Kagbadouno MS, Camara M, Rouamba J, Rayaisse JB, Traoré IS, Camara O, Onikoyamou MF, Courtin F, Ravel S, De Meeûs T, Bucheton B, Jamonneau V, Solano P (2012) Epidemiology of sleeping sickness in boffa (Guinea): where are the trypanosomes? PLoS Neglected Tropical Diseases, 6, e1949. https://doi.org/10.1371/journal.pntd.0001949 Moore A, Richer M, Enrile M, Losio E, Roberts J, Levy D. Resurgence of sleeping sickness in Tambura County, Sudan. Am J Trop Med Hyg. 1999 Aug;61(2):315-8. https://doi.org/10.4269/ajtmh.1999.61.315 Sah R, Mohanty A, Rohilla R, Padhi BK. A resurgence of Sleeping sickness amidst the COVID-19 pandemic: Correspondence. Int J Surg Open. 2023 Apr;53:100604. https://doi.org/10.1016/j.ijso.2023.100604 Welburn SC, Maudlin I. Priorities for the elimination of sleeping sickness. Adv Parasitol. 2012;79:299-337. https://doi.org/10.1016/B978-0-12-398457-9.00004-4 World Health Organization, 2021. Ending the neglect to attain the Sustainable Development Goals: a road map for neglected tropical diseases 2021–2030. World Health Organization, Geneva, Switzerland. ISBN: 978 92 4 001035 2. 196p. World Health Organization, 2023a. Trypanosomiasis, human African (sleeping sickness): key facts. Accessed at https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness) on February 19, 2023. World Health Organization, 2023b. Human African Trypanosomiasis, (sleeping sickness): the global health observatory. Accessed at https://www.who.int/data/gho/data/themes/topics/human-african-trypanosomiasis on February 19, 2023. | Population genetics of *Glossina palpalis* gambiensis in the sleeping sickness focus of Boffa (Guinea) before and after eight years of vector control: no effect of control despite a significant decrease of human exposure to the disease | Moise S. Kagbadouno, Modou Séré, Adeline Ségard, Abdoulaye Dansy Camara, Mamadou Camara, Bruno Bucheton, Jean-Mathieu Bart, Fabrice Courtin, Thierry de Meeûs, Sophie Ravel | <p style="text-align: justify;">Human African trypanosomosis (HAT), also known as sleeping sickness, is still a major concern in endemic countries. Its cyclical vector are biting insects of the genus Glossina or tsetse flies. In Guinea, the mangro... | Disease Ecology/Evolution, Ecology of hosts, infectious agents, or vectors, Evolution of hosts, infectious agents, or vectors, Parasites, Population genetics of hosts, infectious agents, or vectors | Hugues Nana Djeunga | 2023-07-29 13:24:52 | View |
MANAGING BOARD
Jorge Amich
Christine Chevillon
Fabrice Courtin
Christine Coustau
Thierry De Meeûs
Heather R. Jordan
Karl-Heinz Kogel
Yannick Moret
Thomas Pollet
Benjamin Roche
Benjamin Rosenthal
Bashir Salim
Lucy Weinert










